Contrasting Influences of Geographic Range and Distribution of Populations on Patterns of Genetic Diversity in Two Sympatric Pilbara Acacias
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Additional Information
Current Survey Introduced Flora Records Vegetation Condition *Acetosa vesicaria Excellent 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 mE 535,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 536,000 mE 536,000 537,000 mE 537,000 534,000 mE 534,000 mE 535,000 534,000 mE 534,000 -

Genetic Diversity and Differentiation of Invasive Acacia Longifolia in Portugal
Web Ecol., 18, 91–103, 2018 https://doi.org/10.5194/we-18-91-2018 © Author(s) 2018. This work is distributed under the Creative Commons Attribution 4.0 License. Genetic diversity and differentiation of invasive Acacia longifolia in Portugal Sara Vicente1,2, Cristina Máguas2, and Helena Trindade1 1Centro de Estudos do Ambiente e do Mar (CESAM), Centro de Biotecnologia Vegetal (CBV), Faculdade de Ciências da Universidade de Lisboa, Lisbon, 1749-016, Portugal 2Center for Ecology, Evolution and Environmental Changes (cE3c), Faculdade de Ciências da Universidade de Lisboa, Lisbon, 1749-016, Portugal Correspondence: Helena Trindade ([email protected]) Received: 30 January 2018 – Revised: 27 April 2018 – Accepted: 4 May 2018 – Published: 24 May 2018 Abstract. Acacia longifolia is an aggressive invader worldwide. This species was brought to Portugal by the forestry services and is now found throughout the country with very strong ecological and social impacts. Al- though several ecological and physiological studies have been carried out, molecular studies in this species are sparse. Particularly, genetic variability evaluation in invasive ranges clearly deserves more attention. The aim of this study was to evaluate the genetic diversity and the genetic differentiation among populations of the alien in- vasive A. longifolia under different Mediterranean conditions. For that we studied three sandy dune populations along the Portuguese coast: Osso da Baleia (mesomediterranean), Pinheiro da Cruz and Vila Nova de Milfontes (termomediterranean). All sampled acacia plants were located underneath a pine forest, with the exception of the latter case study, where we also sampled in a diversified habitat conditions associated with the margins of agriculture fields. -
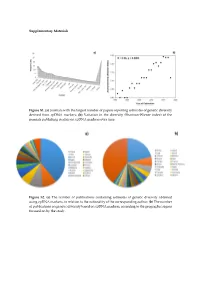
(A) Journals with the Largest Number of Papers Reporting Estimates Of
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

To Name Those Lost: Assessing Extinction Likelihood in the Australian Vascular Flora J.L
To name those lost: assessing extinction likelihood in the Australian vascular flora J.L. SILCOCK, A.R. FIELD, N.G. WALSH and R.J. FENSHAM SUPPLEMENTARY TABLE 1 Presumed extinct plant taxa in Australia that are considered taxonomically suspect, or whose occurrence in Australia is considered dubious. These require clarification, and their extinction likelihood is not assessed here. Taxa are sorted alphabetically by family, then species. No. of Species EPBC1 Last collections References and/or pers. (Family) (State)2 Notes on taxonomy or occurrence State Bioregion/s collected (populations) comms Trianthema cypseleoides Sydney (Aizoaceae) X (X) Known only from type collection; taxonomy needs to be resolved prior to targeted surveys being conducted NSW Basin 1839 1 (1) Steve Douglas Frankenia decurrens (Frankeniaceae) X (X) Very close to F.cinerea and F.brachyphylla; requires taxonomic work to determine if it is a good taxon WA Warren 1850 1 (1) Robinson & Coates (1995) Didymoglossum exiguum Also occurs in India, Sri Lanka, Thailand, Malay Peninsula; known only from type collection in Australia by Domin; specimen exists, but Field & Renner (2019); Ashley (Hymenophyllaceae) X (X) can't rule out the possibility that Domin mislabelled some of these ferns from Bellenden Ker as they have never been found again. QLD Wet Tropics 1909 1 (1) Field Hymenophyllum lobbii Domin specimen in Prague; widespread in other countries; was apparently common and good precision record, so should have been Field & Renner (2019); Ashley (Hymenophyllaceae) X (X) refound by now if present QLD Wet Tropics 1909 1 (1) Field Avon Wheatbelt; Esperance Known from four collections between 1844 and 1892; in her unpublished conspectus of Hemigenia, Barbara Rye included H. -

Annual Report 2008-2009 Annual Report 0
Department of Environment and Conservation and Environment of Department Department of Environment and Conservation 2008-2009 Annual Report 2008-2009 Annual Report Annual 2008-2009 0 ' "p 2009195 E R N M O V E G N T E O H T F W A E I S L T A E R R N A U S T Acknowledgments This report was prepared by the Corporate Communications Branch, Department of Environment and Conservation. For more information contact: Department of Environment and Conservation Level 4 The Atrium 168 St Georges Terrace Perth WA 6000 Locked Bag 104 Bentley Delivery Centre Western Australia 6983 Telephone (08) 6364 6500 Facsimile (08) 6364 6520 Recommended reference The recommended reference for this publication is: Department of Environment and Conservation 2008–2009 Annual Report, Department of Environment and Conservation, 2009. We welcome your feedback A publication feedback form can be found at the back of this publication, or online at www.dec.wa.gov.au. ISSN 1835-1131 (Print) ISSN 1835-114X (Online) 8 September 2009 Letter to THE MINISter Back Contents Forward Hon Donna Faragher MLC Minister for Environment In accordance with section 63 of the Financial Management Act 2006, I have pleasure in submitting for presentation to Parliament the Annual Report of the Department of Environment and Conservation for the period 1 July 2008 to 30 June 2009. This report has been prepared in accordance with provisions of the Financial Management Act 2006. Keiran McNamara Director General DEPARTMENT OF ENVIRONMENT AND CONSERVATION 2008–2009 ANNUAL REPORT 3 DIRECTOR GENERAL’S FOREWORD Back Contents Forward This is the third annual report of the Department of Environment and Conservation since it was created through the merger of the former Department of Environment and Department of Conservation and Land Management. -

SCIENCE COMMUNICATIONS No
SCIENCE COMMUNICATIONS No. 33 (July 2013) Science Division Mission: The provision of up-to-date and scientifically sound information to uphold effective evidence -based conservation of biodiversity and sustainable natural resource management in Western Australia http://intranet/science/ Science Communications lists titles of publications, reports etc prepared by Bachelet E, Fouqué P, Han C, Gould A, Albrow Science Division staff. This issue is for the MD, Beaulieu J-P et al. [Martin R, Williams A] period February-July 2013. (2012). A brown dwarf orbiting an M-dwarf: MOA If you require a copy of a submitted paper 2009-BLG-411L. Astronomy and Astrophysics please contact the author. For all other titles 547, 1-12 please contact [email protected] Bachelet E, Shin I-G, Han C, Fouqué P, Gould A, Margaret Byrne Menzies JW et al. [Martin R, Williams A] (2012). Director MOA 2010-BLG-477Lb: constraining the mass of Science Division a microlensing planet from microlensing parallax, orbital motion and detection of blended light. Astrophysical Journal 754, 1-17 \\DEC- Journal Publications 93sxb2s\InmagicDocs\DEConly\C19271.pdf Abbott I (2012). Depletion of the avifauna of the Beardsley AP, Hazelton BJ, Morales MF, Arcus W, North Island of New Zealand: an 1840s Barnes D, Bernardi G et al. [Williams A] (2013). perspective. Memoirs of the Nuttall The EoR sensitivity of the Murchison Widefield Ornithological Club 18, 51-88 \\DEC- Array. Monthly Notices of the Royal 93sxb2s\InmagicDocs\DEConly\C19131.pdf Astronomical Society: Letters 429, 5-9 \\DEC- 93sxb2s\InmagicDocs\DEConly\C19259.pdf Abbott I (2013). Extending the application of Aboriginal names to Australian biota: Dasyurus Beardsley AP, Hazelton BJ, Morales MF, Capallo (Marsupialia: Dasyuridae) species. -

A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, Near Tom Price
AA VVeeggeettaattiioonn aanndd FFlloorraa SSuurrvveeyy ooff tthhee BBrroocckkmmaann SSyynncclliinnee 44 PPrroojjeecctt AArreeaa,, nneeaarr TToomm PPrriiccee Prepared for Hamersley Iron Pty Ltd Prepared by JJuulllyy 22000055 Biota Environmental Sciences Pty Ltd A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, near Tom Price © Biota Environmental Sciences Pty Ltd 2005 ABN 49 092 687 119 14 View Street North Perth Western Australia 6006 Ph: (08) 9328 1900 Fax: (08) 9328 6138 Project No.: 271 Prepared by: Michi Maier Checked by: Garth Humphreys This document has been prepared to the requirements of the client identified on the cover page and no representation is made to any third party. It may be cited for the purposes of scientific research or other fair use, but it may not be reproduced or distributed to any third party by any physical or electronic means without the express permission of the client for whom it was prepared or Biota Environmental Sciences Pty Ltd. Cube:Current:271 (Brockman 4 Biological):Doc:flora:flora_survey_7.doc 2 A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, near Tom Price A Vegetation and Flora Survey of the Brockman Syncline 4 Project Area, near Tom Price Contents 1.0 Summary 6 1.1 Background 6 1.2 Vegetation 6 1.3 Flora 7 1.4 Management Recommendations 7 2.0 Introduction 9 2.1 Background to the BS4 Project and Location of the Project Area 9 2.2 Scope and Objectives of this Study 9 2.3 Purpose of this Report 12 2.4 Existing Environment 12 3.0 Methodology 18 3.1 Desktop -

Brockman 4 Camps Vegetation and Flora Survey
Biota (n): The living creatures of an area; the flora and fauna together 21 August 2012 Peter Royce Principal Advisor - Environmental Approvals Approvals Division Rio Tinto Central Park 152-158 St George’s Terrace Perth WA 6000 (via email) Dear Peter Brockman 4 Camps Vegetation and Flora Survey Biota Environmental Sciences (Biota) was commissioned by Rio Tinto Pty Ltd (Rio Tinto) to conduct a vegetation and flora survey for the Brockman Camps area (hereafter referred to as the “study area”), located east of the operational BS4 mine and immediately adjacent to the existing accommodation village. This brief report presents the results obtained during that field work. Scope and Objectives The potential development site (defined by the boundaries of the study area) around the Brockman Camps extends over an area of 180 ha, of which 53 ha has been cleared or is deemed as extensively disturbed. Prior to this clearing, a survey conducted by Biota in early December 2006 and late January 2007 recorded no flora of conservation significance in this area (Biota 2007). Vegetation over about 30 ha of the remaining area was previously mapped as part of the Brockman Syncline 4 (BS4) project (Biota 2006). The botanical field survey was therefore conducted over the remaining 97 ha of the study area (adjoining the previously surveyed BS4 study area). This survey was undertaken in accordance with the Guidance Statement No. 51 "Terrestrial Flora and Vegetation Surveys for Environmental Impact Assessment in Western Australia" (EPA 2004). The scope and objectives -

An Inventory of Rangelands in Part of the Broome Shire, Western Australia
Research Library Technical Bulletins Research Publications 2005 An inventory of rangelands in part of the Broome Shire, Western Australia W E. Cotching Follow this and additional works at: https://researchlibrary.agric.wa.gov.au/tech_bull Part of the Agricultural and Resource Economics Commons, Agricultural Economics Commons, Agricultural Science Commons, Desert Ecology Commons, Environmental Education Commons, Environmental Health Commons, Environmental Indicators and Impact Assessment Commons, Environmental Monitoring Commons, Geology Commons, Geomorphology Commons, Natural Resource Economics Commons, Natural Resources and Conservation Commons, Natural Resources Management and Policy Commons, Physical and Environmental Geography Commons, Soil Science Commons, Sustainability Commons, Systems Biology Commons, and the Terrestrial and Aquatic Ecology Commons Recommended Citation Cotching, W E. (2005), An inventory of rangelands in part of the Broome Shire, Western Australia. Department of Primary Industries and Regional Development, Western Australia, Perth. Technical Bulletin 93. This technical bulletin is brought to you for free and open access by the Research Publications at Research Library. It has been accepted for inclusion in Technical Bulletins by an authorized administrator of Research Library. For more information, please contact [email protected]. An inventory of rangelands in part of the Broome Shire, Western Australia W.E. Cotching An inventory of rangelands in part of the Broome Shire, Western Australia By: W.E. Cotching Technical Bulletin No. 93 June 2005 Department of Agriculture 3 Baron-Hay Court SOUTH PERTH 6151 Western Australia ISSN 0083-8675 © State of Western Australia, 2005 Acknowledgments This document was first prepared in 1990, but remained unpublished. It was recompiled in 1998 as part of the Natural Resource Assessment Group ‘Value Adding’ project, which seeks to consolidate and document in a database, a vast amount of published and (especially) unpublished land resource information. -

SCIENCE COMMUNICATIONS No
SCIENCE COMMUNICATIONS No. 34 (January 2014) Science & Conservation Division Mission: The provision of up-to-date and scientifically sound information to uphold effective evidence-based conservation of biodiversity and sustainable natural resource management in Western Australia Science Communications lists titles of Davison EM, McGurk LE, Bougher NL, Syme K, publications, reports etc prepared by Science Watkin ELJ (2013). Amanita lesueuri and and Conservation Division staff. This issue is for Amanita wadjukorum (Basidiomycota), two new the period August 2013.-January 2014 species from Western Australia, and an If you require a copy of a submitted paper please expanded description of Amanita fibrillopes. contact the author. For all other titles please Nuytsia 23, 589-606 \\KENS-file-001 contact [email protected] \Inmagic\FullTextFiles/080057-23.031.pdf Gosper CR, Prober SM, Yates CJ (2013). Multi- Margaret Byrne century changes in vegetation structure and fuel Director availability in fire-sensitive eucalypt woodlands. Science & Conservation Division Forest Ecology and Management 310, 102-109 \\KENS-file-001\Inmagic\StaffOnly/C19292.pdf Journal Publications Gosper C, Prober S, Yates C (2013). The role of Barrett RL, Lowrie A (2013). Typification and fire in plant conservation in wheatbelt remnants. application of names in Drosera section Australasian Plant Conservation 22, 21-22 Arachnopus (Droseraceae). Nuytsia 23, 527-541 \\KENS-file-001\Inmagic\StaffOnly/C19281.pdf \\KENS-file-001\Inmagic\FullTextFiles/080057- 23.029.pdf Gosper CR, Yates CJ, Prober SM (2013). Floristic diversity in fire-sensitive eucalypt woodlands Biggs LJ, Parker CL (2013). Updates to Western show a U-shaped relationship with time since Australia's vascular plant census for 2012. -

The Australian Centre for International Agricultural Research (ACIAR) Was Established in June 1982 by an Act of the Australian Parliament
The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. Its mandate is to help identify agricultural problems in developing countries and to commission collaborative research between Australian and developing country researchers in fields where Australia has a special research competence. Where trade names are used this does not constitute endorsement of nor discrimination against any product by the Centre. ACIAR PROCEEDINGS This series of publications includes the full proceedings of research workshops or symposia organised or supported by ACIAR. Numbers in this series are distrib uted internationally to selected individuals and scientific institutions. Previous numbers in the series are listed on the inside back cover. © Australian Centre for International Agricultural Research G.P.O. Box 1571, Canberra, A.C.T. 2601 Turnbull, John W. 1987. Australian acacias in developing countries: proceedings of an international workshop held at the Forestry Training Centre, Gympie, Qld., Australia, 4-7 August 1986. ACIAR Proceedings No. 16, 196 p. ISBN 0 949511 269 Typeset and laid out by Union Offset Co. Pty Ltd, Fyshwick, A.C.T. Printed by Brown Prior Anderson Pty Ltd, 5 Evans Street Burwood Victoria 3125 Australian Acacias in Developing Countries Proceedings of an international workshop held at the Forestry Training Centre, Gympie, Qld., Australia, 4-7 August 1986 Editor: John W. Turnbull Workshop Steering Committee: Douglas 1. Boland, CSIRO Division of Forest Research Alan G. Brown, CSIRO Division of Forest Research John W. Turnbull, ACIAR and NFTA Paul Ryan, Queensland Department of Forestry Cosponsors: Australian Centre for International Agricultural Research (ACIAR) Nitrogen Fixing Tree Association (NFTA) CSIRO Division of Forest Research Queensland Department of Forestry Contents Foreword J . -

Case Study of Status and Change in the Rangelands of the Gascoyne – Murchison Region
Case study of status and change in the rangelands of the Gascoyne – Murchison region Report to the Australian Collaborative Rangeland Information System (ACRIS) Management Committee 1998 1998 1998 1994 1998 1999 2003 2003 2003 2000 2002 2002 Ian Watson, Jeff Richardson, Philip Thomas and Damian Shepherd http://www.deh.gov.au/land/management/rangelands/acris/index.html ISBN 0642551847 Copyright and Disclaimers © Commonwealth of Australia 2006 Information contained in this publication may be copied or reproduced for study, research, information or educational purposes, subject to inclusion of an acknowledgment of the source. The views and opinions expressed in this publication are those of the authors and do not necessarily reflect those of the Australian Government or the Minister for the Environment and Heritage. While reasonable efforts have been made to ensure that the contents of this publication are factually correct, the Australian Government does not accept responsibility for the accuracy or completeness of the contents, and shall not be liable for any loss or damage that may be occasioned directly or indirectly through the use of, or reliance on, the contents of this publication. Acknowledgements A large number of people helped the authors compile this report. Gary Bastin, the ACRIS Coordinator, provided much insight, encouragement and advice on what was useful at the national scale. Gary also did a fantastic job of synthesising the mass of disparate information to come out of the pilot project reports and other data sources into a coherent national report. Members of the ACRIS management committee (past and present) provided good ideas, support and encouragement for the set of ACRIS pilot projects.