Required Software Environments in Advance, So That Everyone Can Run the Program Code on Their Own Laptop During the Hands-On Sessions
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Linux on the Road
Linux on the Road Linux with Laptops, Notebooks, PDAs, Mobile Phones and Other Portable Devices Werner Heuser <wehe[AT]tuxmobil.org> Linux Mobile Edition Edition Version 3.22 TuxMobil Berlin Copyright © 2000-2011 Werner Heuser 2011-12-12 Revision History Revision 3.22 2011-12-12 Revised by: wh The address of the opensuse-mobile mailing list has been added, a section power management for graphics cards has been added, a short description of Intel's LinuxPowerTop project has been added, all references to Suspend2 have been changed to TuxOnIce, links to OpenSync and Funambol syncronization packages have been added, some notes about SSDs have been added, many URLs have been checked and some minor improvements have been made. Revision 3.21 2005-11-14 Revised by: wh Some more typos have been fixed. Revision 3.20 2005-11-14 Revised by: wh Some typos have been fixed. Revision 3.19 2005-11-14 Revised by: wh A link to keytouch has been added, minor changes have been made. Revision 3.18 2005-10-10 Revised by: wh Some URLs have been updated, spelling has been corrected, minor changes have been made. Revision 3.17.1 2005-09-28 Revised by: sh A technical and a language review have been performed by Sebastian Henschel. Numerous bugs have been fixed and many URLs have been updated. Revision 3.17 2005-08-28 Revised by: wh Some more tools added to external monitor/projector section, link to Zaurus Development with Damn Small Linux added to cross-compile section, some additions about acoustic management for hard disks added, references to X.org added to X11 sections, link to laptop-mode-tools added, some URLs updated, spelling cleaned, minor changes. -

Lightweight Distros on Test
GROUP TEST LIGHTWEIGHT DISTROS LIGHTWEIGHT DISTROS GROUP TEST Mayank Sharma is on the lookout for distros tailor made to infuse life into his ageing computers. On Test Lightweight distros here has always been a some text editing, and watch some Linux Lite demand for lightweight videos. These users don’t need URL www.linuxliteos.com Talternatives both for the latest multi-core machines VERSION 2.0 individual apps and for complete loaded with several gigabytes of DESKTOP Xfce distributions. But the recent advent RAM or even a dedicated graphics Does the second version of the distro of feature-rich resource-hungry card. However, chances are their does enough to justify its title? software has reinvigorated efforts hardware isn’t supported by the to put those old, otherwise obsolete latest kernel, which keeps dropping WattOS machines to good use. support for older hardware that is URL www.planetwatt.com For a long time the primary no longer in vogue, such as dial-up VERSION R8 migrators to Linux were people modems. Back in 2012, support DESKTOP LXDE, Mate, Openbox who had fallen prey to the easily for the i386 chip was dropped from Has switching the base distro from exploitable nature of proprietary the kernel and some distros, like Ubuntu to Debian made any difference? operating systems. Of late though CentOS, have gone one step ahead we’re getting a whole new set of and dropped support for the 32-bit SparkyLinux users who come along with their architecture entirely. healthy and functional computers URL www.sparkylinux.org that just can’t power the newer VERSION 3.5 New life DESKTOP LXDE, Mate, Xfce and others release of Windows. -

Introduction to Fmxlinux Delphi's Firemonkey For
Introduction to FmxLinux Delphi’s FireMonkey for Linux Solution Jim McKeeth Embarcadero Technologies [email protected] Chief Developer Advocate & Engineer For quality purposes, all lines except the presenter are muted IT’S OK TO ASK QUESTIONS! Use the Q&A Panel on the Right This webinar is being recorded for future playback. Recordings will be available on Embarcadero’s YouTube channel Your Presenter: Jim McKeeth Embarcadero Technologies [email protected] | @JimMcKeeth Chief Developer Advocate & Engineer Agenda • Overview • Installation • Supported platforms • PAServer • SDK & Packages • Usage • UI Elements • Samples • Database Access FireDAC • Migrating from Windows VCL • midaconverter.com • 3rd Party Support • Broadway Web Why FMX on Linux? • Education - Save money on Windows licenses • Kiosk or Point of Sale - Single purpose computers with locked down user interfaces • Security - Linux offers more security options • IoT & Industrial Automation - Add user interfaces for integrated systems • Federal Government - Many govt systems require Linux support • Choice - Now you can, so might as well! Delphi for Linux History • 1999 Kylix: aka Delphi for Linux, introduced • It was a port of the IDE to Linux • Linux x86 32-bit compiler • Used the Trolltech QT widget library • 2002 Kylix 3 was the last update to Kylix • 2017 Delphi 10.2 “Tokyo” introduced Delphi for x86 64-bit Linux • IDE runs on Windows, cross compiles to Linux via the PAServer • Designed for server side development - no desktop widget GUI library • 2017 Eugene -

Linux Lite 1.0.2 – Basic Computer And
computer literacy movement construction e - learning of a modern ITdesk.info – society project of computer e-education with an open access - digital literacy handbook human rights e - inclusion to education and information open access Linux Lite 1.0.2 – Basic computer and file managing HANDBOOK Author: Melani Max ITdesk.info – project of computer e-education with open access Original: Author: Gorana Čelebić Subtitle: Osnovno korištenje računala i upravljanje datotekama – Windows 7, priručnik ISBN: 978-953-56758-1-5 Publisher: Otvoreno društvo za razmjenu ideja (ODRAZI), Zagreb Expert review: EdukaCentar, ustanova za obrazovanje, Zagreb Proofreading: Ema Čišić Place and year of publication: Zagreb, 2011. Adjusted for the Linux Lite operating system by: Author: Melani Max Main title: ITdesk.info – projekt računalne e-edukacije sa slobodnim pristupom Subtitle: Linux Lite 1.0.2 - Basic computer and file managing, handbook Translation: Una Šijan Proof-reading: Manuela Sánchez Expert review: Kundan Singh Publisher: Otvoreno društvo za razmjenu ideja (ODRAZI), Zagreb ISBN: 978-953-56758-1-5 Place and year of publication: Zagreb, 2013. Copyright: Feel free to copy, print and re-distribute the whole or a part of this publication, including for the educational purposes, whether public or private educational organizations, but exclusively in non- commercial purposes (therefore, without charging the end users of publication) and with a reference to the source (source: www.ITdesk.info – computer e-education project with a free access). Derivative works without prior approval of the copywright holder (Open Society for the Exchange of Ideas-ODRAZI) are not permitted. Contact for permission for derivative works: [email protected]. ITdesk.info – project of computer e-education with open access PREFACE Today’s society is marked by a sudden growth and development of information technology (IT) field, which has resulted in society being greatly dependent, in a broader sense, on the knowledge and competence of the people working in the IT sector. -
Opensuse 32 Bit Iso Download Opensuse 32 Bit Iso Download
opensuse 32 bit iso download Opensuse 32 bit iso download. Completing the CAPTCHA proves you are a human and gives you temporary access to the web property. What can I do to prevent this in the future? If you are on a personal connection, like at home, you can run an anti-virus scan on your device to make sure it is not infected with malware. If you are at an office or shared network, you can ask the network administrator to run a scan across the network looking for misconfigured or infected devices. Another way to prevent getting this page in the future is to use Privacy Pass. You may need to download version 2.0 now from the Chrome Web Store. Cloudflare Ray ID: 669f49e8e8a97b7f • Your IP : 188.246.226.140 • Performance & security by Cloudflare. Opensuse 32 bit iso download. Completing the CAPTCHA proves you are a human and gives you temporary access to the web property. What can I do to prevent this in the future? If you are on a personal connection, like at home, you can run an anti-virus scan on your device to make sure it is not infected with malware. If you are at an office or shared network, you can ask the network administrator to run a scan across the network looking for misconfigured or infected devices. Another way to prevent getting this page in the future is to use Privacy Pass. You may need to download version 2.0 now from the Chrome Web Store. Cloudflare Ray ID: 669f49e96aadf14e • Your IP : 188.246.226.140 • Performance & security by Cloudflare. -

Delphi's Firemonkey for Linux Solution
Introduction to FMXLinux Delphi’s FireMonkey for Linux Solution Jim McKeeth Embarcadero Technologies [email protected] Chief Developer Advocate & Engineer Slides, replay and more https://embt.co/FMXLinuxIntro Your Presenter: Jim McKeeth Embarcadero Technologies [email protected] | @JimMcKeeth Chief Developer Advocate & Engineer Agenda • Overview • Installation • Supported platforms • PAServer • SDK & Packages • Usage • UI Elements • Samples • Database Access FireDAC • Migrating from Windows VCL • midaconverter.com • 3rd Party Support • Broadway Web Why FMX on Linux? • Education - Save money on Windows licenses • Kiosk or Point of Sale - Single purpose computers with locked down user interfaces • Security - Linux offers more security options • IoT & Industrial Automation - Add user interfaces for integrated systems • Federal Government - Many govt systems require Linux support • Choice - Now you can, so might as well! Delphi for Linux History • 1999 Kylix: aka Delphi for Linux, introduced • It was a port of the IDE to Linux • Linux x86 32-bit compiler • Used the Trolltech QT widget library • 2002 Kylix 3 was the last update to Kylix • 2017 Delphi 10.2 “Tokyo” introduced Delphi for x86 64-bit Linux • IDE runs on Windows, cross compiles to Linux via the PAServer • Designed for server side development - no desktop widget GUI library • 2017 Eugene Kryukov of KSDev release FMXLinux • Eugene was one of the original architects of FireMonkey • A modification of FireMonkey, bringing FMX to Linux • 2019 Embarcadero includes FMXLinux -

Lexmark Xc6152dtfe
Lexmark XC6152dtfe The Lexmark XC6152 workgroup colour A4 MFP prints up to 50 pages per minute and has business-class features like pre- installed software solutions and an inline staple finisher. Part Number Scanner Type Novell Operating Systems Supported 42K1185 Flatbed scanner with ADF Novell NetWare 6.5 Open Enterprise Server, Print Technology Scan Technology Novell Open Enterprise Server 2 on SUSE Color Laser CCD Linux Enterprise Server 10 SP4, Novell Open Enterprise Server 11 on SUSE Linux Enterprise Function Optical Scanning Resolution Server 11 Colour Copying, Colour Faxing, Colour 1200 X 600 ppi UNIX Operating Systems Supported Printing, Colour Scanning, Colour Network Scan File Formats (Send as) Scanning Sun Solaris x86 10, 11, Sun Solaris SPARC 10, JPEG, JPG, PDF, Secure PDF, TIFF, XPS, 11, IBM AIX 6.1, 7.1 Workgroup Size Archival PDF (A-1a, A-1b), Searchable PDF Medium-Large Workgroup (optional OCR with 1GB RAM and hard disk Included Paper Handling 550-Sheet Input, 100-Sheet Multipurpose Touch Screen Display required), Highly compressed PDF (1GB RAM and hard disk required) Feeder, Integrated Duplex, 2 x 550-Sheet Yes Tray, Inline Stapler, 300-Sheet Output Bin Display Scan Destinations e-mail, FTP, USB Flash drive, USB or Network Optional Paper Handling Lexmark e-Task 17.8 cm (7-inch) colour touch 550-Sheet Tray, 2200-Sheet Tray screen connected computer, Lexmark Document Solutions Suite (LDSS) Paper Input Capacity, Standard Print Speed (A4, Black) Modem Speed Up to 1,750 pages 75 gsm bond Up to 50 ppm6 ITU T.30, V.34 Half-Duplex, -

Free Download Linux
free download linux 10 Reasons: Why should we use Linux? Bored using windows or any other server in your computing experience? Linux has been designed to offer you a smooth and effective experience. Whether you want to use it for business or any other home operations, Linux offers the most reliable system to adopt. Linux has become one of the most adopted systems by many users in and around the world. Most of the world’s largest data centers across the globe use Linux. There are various reasons that answer why should we use Linux, keep on reading. Why Should we use Linux? 1. Linux is free of charge and in case you still doubt, then know that it is. In case you want to get rid of all the restrictions in terms of patents of any other form of restriction, then this would be the best switch for you. Linux can be edited in terms of the source code by most of the innovative business owners. It is, therefore, the best system to be adopted by a large number of business owners who want to simplify their business operations. 2. Linux is more secure than any other server. Linux is a dependable server that offers excellent security to the user. Linux community offers the security fixes for any technical or security threat that encounters. Moreover, Linux users also offer regular kernel updates and regular security patches. In this regard, it is not mandatory for you to have an anti-virus program for your system. With Linux, you can save a lot of dollars you could have spent buying an antivirus program. -
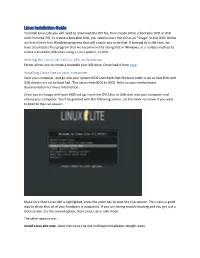
Linux Installation Guide to Install Linux Lite You Will Need to Download the ISO File, Then Create Either a Bootable DVD Or USB Stick from the ISO
Linux Installation Guide To install Linux Lite you will need to download the ISO file, then create either a bootable DVD or USB stick from the ISO. To create a bootable DVD, you need to burn the ISO as an "image" to the DVD. Below are listed three free Windows programs that will enable you to do that. If burning to a USB stick, we have also listed a free program that we recommend for doing that in Windows; or a reliable method to create a bootable USB when using a Linux system, or OSX. Writing the Linux Lite ISO to USB on Windows Etcher allows you to create a bootable Live USB drive. Download it from here. Installing Linux Lite to your computer Start your computer, and go into your system BIOS and check that the boot order is set so that DVD and USB devices are set to boot first. This varies from BIOS to BIOS. Refer to your motherboard documentation for more information. Once you are happy with your BIOS set up, insert the DVD disc or USB stick into your computer and reboot your computer. You'll be greeted with the following screen. Let the timer run down if you want to boot to the Live session. Make sure Start Linux Lite is highlighted, press the enter key to boot the Live session. This is also a good way to check that all of your hardware is supported. If you are having trouble booting and you get just a black screen, try the second option, Start Linux Lite in safe mode. -
Ubuntu 14.04 Respins
FEATURE UBUNTU 14.04 RESPINS UBUNTU 14.04 RESPINS Love Ubuntu, but hate feature X? Mayank Sharma shows you over a dozen different spins that make Ubuntu enjoyable again. veryone has their favourite Ubuntu feature the distro’s installer, which is one of the best tools they dislike. Privacy-conscious users loathe for the job, and Canonical is also responsible for Ethe Lenses feature. Desktop users get unleashing ideas, such as the Software Centre irritated with the window controls on the wrong and Ubuntu One, that have helped change the side of the window and the overlay scrollbars. And perception of Linux as a desktop OS. Gnome 2 users can’t bring themselves to forgive As the distro unveils its next Long Term Support the distro for ruining their usability with the Unity release, we introduce you to some of the distros desktop and the lack of customisation options. that have stood on the shoulder of giants, remixing But for every feature that’s rubbed users the the best of Ubuntu with the much wider open wrong way, Ubuntu has contributed several useful source community to create several refreshing new ones that’ve benefitted the community. There’s flavours of what Linux can be. 18 www.linuxvoice.com UBUNTU 14.04 RESPINS FEATURE Ultimate Edition Do you want one with everything? The aptly-named Ultimate Edition distro is Avidemux video editor and Blender for 3D loaded with tons of Free and proprietary modelling. For package management, the software. The latest version is based on distro bundles the Ubuntu Software Centre Ubuntu 14.04 and uses the Mate desktop along with the Synaptic package manager. -
The Following Distributions Match Your Criteria (Sorted by Popularity): 1. Linux Mint (1) Linux Mint Is an Ubuntu-Based Distribu
The following distributions match your criteria (sorted by popularity): 1. Linux Mint (1) Linux Mint is an Ubuntu-based distribution whose goal is to provide a more complete out-of-the-box experience by including browser plugins, media codecs, support for DVD playback, Java and other components. It also adds a custom desktop and menus, several unique configuration tools, and a web-based package installation interface. Linux Mint is compatible with Ubuntu software repositories. 2. Mageia (2) Mageia is a fork of Mandriva Linux formed in September 2010 by former employees and contributors to the popular French Linux distribution. Unlike Mandriva, which is a commercial entity, the Mageia project is a community project and a non-profit organisation whose goal is to develop a free Linux-based operating system. 3. Ubuntu (3) Ubuntu is a complete desktop Linux operating system, freely available with both community and professional support. The Ubuntu community is built on the ideas enshrined in the Ubuntu Manifesto: that software should be available free of charge, that software tools should be usable by people in their local language and despite any disabilities, and that people should have the freedom to customise and alter their software in whatever way they see fit. "Ubuntu" is an ancient African word, meaning "humanity to others". The Ubuntu distribution brings the spirit of Ubuntu to the software world. 4. Fedora (4) The Fedora Project is an openly-developed project designed by Red Hat, open for general participation, led by a meritocracy, following a set of project objectives. The goal of The Fedora Project is to work with the Linux community to build a complete, general purpose operating system exclusively from open source software. -
Ubuntu: Review of LTS 16.04.02, Latest Developments, and Linux Lite 3.6
Published on Tux Machines (http://www.tuxmachines.org) Home > content > Ubuntu: Review of LTS 16.04.02, Latest Developments, and Linux Lite 3.6 Ubuntu: Review of LTS 16.04.02, Latest Developments, and Linux Lite 3.6 By Roy Schestowitz Created 05/09/2017 - 10:29am Submitted by Roy Schestowitz on Tuesday 5th of September 2017 10:29:31 AM Filed under Ubuntu [1] Review: Ubuntu LTS 16.04.02 has a long shelf-life [2] Controlling snap releases with channels, tracks and branches ? Part 1 [3] Ever since snaps were introduced, a crucial part of their feature set has been the ability to release a snap on a particular ?channel? indicating how stable or production-ready it is. The well-known channel names stable, candidate, beta and edge indicate a snap?s stability, according to the developer, and users are then empowered to choose the level of risk they are prepared to accept when installing a snap. From a developer?s point of view, it makes sense to release new changes on edge, which will presumably have a small number of users willing to accept some instability in exchange for bleeding-edge features (but also, tacitly, a disposition to report problems), and as any rough edges are taken care of, release the updated snap to beta, candidate and finally to stable once it?s deemed adequate for anyone to use. Ubuntu 17.10 (Artful Aardvark) Beta 1 Out for Opt-In Flavors, Here's What's New [4] On the last day of August, the upcoming Ubuntu 17.10 (Artful Aardvark) operating system entered Beta stages of development, but only for some of the official flavors, including Kubuntu, Xubuntu, Lubuntu, Ubuntu Studio, Ubuntu MATE, Ubuntu Kylin, and Ubuntu Budgie.