Endostatin and Irradiation Modifies the Activity of ADAM10 and Neprilysin in Breast Cancer Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Neprilysin Is Required for Angiotensin-(1-7)
Page 1 of 39 Diabetes NEPRILYSIN IS REQUIRED FOR ANGIOTENSIN-(1-7)’S ABILITY TO ENHANCE INSULIN SECRETION VIA ITS PROTEOLYTIC ACTIVITY TO GENERATE ANGIOTENSIN-(1-2) Gurkirat S. Brara, Breanne M. Barrowa, Matthew Watsonb, Ryan Griesbachc, Edwina Chounga, Andrew Welchc, Bela Ruzsicskad, Daniel P. Raleighb, Sakeneh Zraikaa,c aVeterans Affairs Puget Sound Health Care System, Seattle, WA 98108, United States bDepartment of Chemistry, Stony Brook University, Stony Brook, NY 11794, United States cDivision of Metabolism, Endocrinology and Nutrition, Department of Medicine, University of Washington, Seattle, WA 98195, United States dInstitute for Chemical Biology and Drug Discovery, Stony Brook University, Stony Brook, NY 11794, United States Short Title: Angiotensin-(1-7) and insulin secretion Word count: 3997; Figure count: 8 main (plus 3 Online Suppl.); Table count: 1 Online Suppl. Correspondence to: Sakeneh Zraika, PhD 1660 South Columbian Way (151) Seattle, WA, United States Tel: 206-768-5391 / Fax: 206-764-2164 Email: [email protected] 1 Diabetes Publish Ahead of Print, published online May 30, 2017 Diabetes Page 2 of 39 ABSTRACT Recent work has renewed interest in therapies targeting the renin-angiotensin system (RAS) to improve β-cell function in type 2 diabetes. Studies show that generation of angiotensin-(1-7) by angiotensin converting enzyme 2 (ACE2) and its binding to the Mas receptor (MasR) improves glucose homeostasis, partly by enhancing glucose-stimulated insulin secretion (GSIS). Thus, islet ACE2 upregulation is viewed as a desirable therapeutic goal. Here, we show that although endogenous islet ACE2 expression is sparse, its inhibition abrogates angiotensin-(1-7)-mediated GSIS. However, a more widely expressed islet peptidase, neprilysin, degrades angiotensin-(1-7) into several peptides. -

View a Copy of This Licence, Visit
Choi et al. BMC Cardiovascular Disorders (2020) 20:360 https://doi.org/10.1186/s12872-020-01636-5 RESEARCH ARTICLE Open Access Soluble neprilysin and long-term clinical outcomes in patients with coronary artery disease undergoing percutaneous coronary intervention: a retrospective cohort study Ik Jun Choi1, Sungmin Lim2* , Youngdeok Hwang3, Dongjae Lee1, Won Jik Lee1, Kwan Yong Lee1, Mi-Jeong Kim1 and Doo Soo Jeon1 Abstract Background: Neprilysin has an essential role in regulating fluid balance and vascular resistance, and neprilysin inhibitors have shown beneficial effects in patients with heart failure. However, the potential predictive value of neprilysin levels as a biomarker for cardiovascular risk remains unclear. The aim of this study was to assess the prognostic value of soluble neprilysin (sNEP) levels in patients with ischemic heart disease. Methods: Neprilysin levels were measured in 694 consecutive patients with coronary artery disease (CAD) undergoing percutaneous coronary intervention (PCI). These patients were classified into two groups according to their serum levels of neprilysin and categorized into the lower neprilysin group (n = 348) and the higher neprilysin group (n = 346). The primary clinical endpoint was all-cause mortality, and the secondary endpoint was a composite of major adverse cardiac events (MACE). Results: The median sNEP level was 76.0 pg/ml. The median sNEP levels were higher in patients with left ventricular ejection fraction (LVEF) ≥40% (77.6 pg/ml, interquartile range 46.6–141.3) than in those with LVEF < 40% (70.0 pg/ml, interquartile range 47.1–100.6; P = 0.032). Among all patients, each clinical outcome and MACE did not differ significantly according to the groups divided into median, tertile, or quartile of sNEP levels during a median follow-up of 28.4 months. -

Stromal CD10 Expression in Breast Cancer Correlates with Tumor Invasion and Cancer Stem Cell Phenotype
Louhichi et al. BMC Cancer (2018) 18:49 DOI 10.1186/s12885-017-3951-8 RESEARCH ARTICLE Open Access Stromal CD10 expression in breast cancer correlates with tumor invasion and cancer stem cell phenotype Tahani Louhichi, Hanene Saad, Myriam Ben Dhiab, Sonia Ziadi and Mounir Trimeche* Abstract Background: Previous investigations have indicated that CD10 is associated with biological aggressivity in human cancers, but the use of this marker for diagnosis and prognosis is more complex. The aim of this study was to evaluate the expression of CD10 in breast cancer and its association with the clinicopathological features. In addition, we investigated whether a relationship exists between CD10 expression and cancer stem cells. Methods: CD10 expression was examined by the immunohistochemistry in a series of 133 invasive breast carcinoma cases. Results were correlated to several clinicopathological parameters. Cancer stem cell phenotype was assessed by the immunohistochemical analysis of CD44 and ALDH1. Results: Significant CD10 expression was found in the fusiform stromal cells in 19.5% of the cases and in the neoplastic cells in 7% of the cases. The stromal CD10 positivity was more frequently found in tumors with lymph node metastasis (p = 0.01) and a high histological grade (p = 0.01). However, CD10 expression by the neoplastic cells correlates with a high histological grade (p = 0.03) and the absence of estrogen (p = 0.002) as well as progesterone (p = 0.001) receptor expression. We also found that CD10 expression by the stromal cells, but not by the neoplastic cells, correlates significantly with the expression of cancer stem cell markers (CD44+/ALDH1+) (p = 0.002). -

Supporting Online Material
1 2 3 4 5 6 7 Supplementary Information for 8 9 Fractalkine-induced microglial vasoregulation occurs within the retina and is altered early in diabetic 10 retinopathy 11 12 *Samuel A. Mills, *Andrew I. Jobling, *Michael A. Dixon, Bang V. Bui, Kirstan A. Vessey, Joanna A. Phipps, 13 Ursula Greferath, Gene Venables, Vickie H.Y. Wong, Connie H.Y. Wong, Zheng He, Flora Hui, James C. 14 Young, Josh Tonc, Elena Ivanova, Botir T. Sagdullaev, Erica L. Fletcher 15 * Joint first authors 16 17 Corresponding author: 18 Prof. Erica L. Fletcher. Department of Anatomy & Neuroscience. The University of Melbourne, Grattan St, 19 Parkville 3010, Victoria, Australia. 20 Email: [email protected] ; Tel: +61-3-8344-3218; Fax: +61-3-9347-5219 21 22 This PDF file includes: 23 24 Supplementary text 25 Figures S1 to S10 26 Tables S1 to S7 27 Legends for Movies S1 to S2 28 SI References 29 30 Other supplementary materials for this manuscript include the following: 31 32 Movies S1 to S2 33 34 35 36 1 1 Supplementary Information Text 2 Materials and Methods 3 Microglial process movement on retinal vessels 4 Dark agouti rats were anaesthetized, injected intraperitoneally with rhodamine B (Sigma-Aldrich) to label blood 5 vessels and retinal explants established as described in the main text. Retinal microglia were labelled with Iba-1 6 and imaging performed on an inverted confocal microscope (Leica SP5). Baseline images were taken for 10 7 minutes, followed by the addition of PBS (10 minutes) and then either fractalkine or fractalkine + candesartan 8 (10 minutes) using concentrations outlined in the main text. -

3 Cleavage Products of Notch 2/Site and Myelopoiesis by Dysregulating
ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch This information is current as David R. Gibb, Sheinei J. Saleem, Dae-Joong Kang, Mark of October 4, 2021. A. Subler and Daniel H. Conrad J Immunol 2011; 186:4244-4252; Prepublished online 2 March 2011; doi: 10.4049/jimmunol.1003318 http://www.jimmunol.org/content/186/7/4244 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2011/03/02/jimmunol.100331 Material 8.DC1 http://www.jimmunol.org/ References This article cites 45 articles, 16 of which you can access for free at: http://www.jimmunol.org/content/186/7/4244.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on October 4, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2011 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology ADAM10 Overexpression Shifts Lympho- and Myelopoiesis by Dysregulating Site 2/Site 3 Cleavage Products of Notch David R. -

Angiotensin Receptor Neprilysin Inhibition (ARNI) Following Acute Myocardial Infarction: Primary Results of the PARADISE-MI Trial Marc A
Angiotensin Receptor Neprilysin Inhibition (ARNI) Following Acute Myocardial Infarction: Primary Results of the PARADISE-MI Trial Marc A. Pfeffer, MD, PhD Distinguished Dzau Professor of Medicine Harvard Medical School Cardiovascular Division, Brigham and Women’s Hospital for the PARADISE-MI Committees, National Leaders and Investigators SAVE AIRE TRACE Radionuclide Clinical and/or Echocardiographic EF ≤ 40% radiographic signs EF ≤ 35% (1992) of HF (1993) (1995) 0.4 All-Cause Mortality 0.35 0.3 0.25 Placebo ACE-I 0.2 0.15 Placebo: 866/2971 (29.1%) Probability of of Probability Event 0.1 ACE-I: 702/2995 (23.4%) 0.05 OR: 0.74 (0.66–0.83) 0 Years 0 1 2 3 4 ACE-I 2995 2250 1617 892 223 Placebo 2971 2184 1521 853 138 Flather MD, et al. Lancet. 2000;355:1575–1581 Mortality in SAVE, TRACE, AIRE, and VALIANT Favors Active Drug Pfeffer,Pfeffer, McMurray, McMurray, Velazquez, Velazquez, et etal. al. N NEngl Engl J MedJ Med2003;3492003;349 2014 40 Enalapril 1117 32 (n=4212) 914 24 LCZ696 (n=4187) Meier Estimate of Meier Estimate 16 - HR = 0.80 (0.73-0.87) Cumulative (%) Rates Cumulative 8 Kaplan P = 0.0000002 Number needed to treat = 21 0 0 180 360 540 720 900 1080 1260 Patients at Risk Days After Randomization LCZ696 4187 3922 3663 3018 2257 1544 896 249 Enalapril 4212 3883 3579 2922 2123 1488 853 236 McMurray, N Engl J Med. 2014 AMI (0.5-7 days with LVEF ≤40% and/or pulmonary congestion) PLUS any risk enhancer Age ≥70 years Atrial fibrillation eGFR <60 LVEF < 30% Diabetes Killip class ≥III Prior MI STEMI without reperfusion Major Exclusions: Prior HF Clinical instability eGFR <30 Sacubitril/Valsartan Ramipril No run-in Target 97/103 mg BID Target 5 mg BID double-blind -controlled N=2830 active N=2831 Event driven: 711 primary endpoints Median follow-up: 23 months Primary Endpoint: CV death, HF hospitalization, outpatient development of HF Jering, Eur J ACC.21 Secondary Endpoint: CV death or first HF hospitalization Heart Fail. -

ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease
International Journal of Molecular Sciences Review ADAM10 Site-Dependent Biology: Keeping Control of a Pervasive Protease Francesca Tosetti 1,* , Massimo Alessio 2, Alessandro Poggi 1,† and Maria Raffaella Zocchi 3,† 1 Molecular Oncology and Angiogenesis Unit, IRCCS Ospedale Policlinico S. Martino Largo R. Benzi 10, 16132 Genoa, Italy; [email protected] 2 Proteome Biochemistry, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] 3 Division of Immunology, Transplants and Infectious Diseases, IRCCS San Raffaele Scientific Institute, 20132 Milan, Italy; [email protected] * Correspondence: [email protected] † These authors contributed equally to this work as last author. Abstract: Enzymes, once considered static molecular machines acting in defined spatial patterns and sites of action, move to different intra- and extracellular locations, changing their function. This topological regulation revealed a close cross-talk between proteases and signaling events involving post-translational modifications, membrane tyrosine kinase receptors and G-protein coupled recep- tors, motor proteins shuttling cargos in intracellular vesicles, and small-molecule messengers. Here, we highlight recent advances in our knowledge of regulation and function of A Disintegrin And Metalloproteinase (ADAM) endopeptidases at specific subcellular sites, or in multimolecular com- plexes, with a special focus on ADAM10, and tumor necrosis factor-α convertase (TACE/ADAM17), since these two enzymes belong to the same family, share selected substrates and bioactivity. We will discuss some examples of ADAM10 activity modulated by changing partners and subcellular compartmentalization, with the underlying hypothesis that restraining protease activity by spatial Citation: Tosetti, F.; Alessio, M.; segregation is a complex and powerful regulatory tool. -

Cell-Autonomous FLT3L Shedding Via ADAM10 Mediates Conventional Dendritic Cell Development in Mouse Spleen
Cell-autonomous FLT3L shedding via ADAM10 mediates conventional dendritic cell development in mouse spleen Kohei Fujitaa,b,1, Svetoslav Chakarovc,1, Tetsuro Kobayashid, Keiko Sakamotod, Benjamin Voisind, Kaibo Duanc, Taneaki Nakagawaa, Keisuke Horiuchie, Masayuki Amagaib, Florent Ginhouxc, and Keisuke Nagaod,2 aDepartment of Dentistry and Oral Surgery, Keio University School of Medicine, Tokyo 160-8582, Japan; bDepartment of Dermatology, Keio University School of Medicine, Tokyo 160-8582, Japan; cSingapore Immunology Network, Agency for Science, Technology and Research, Biopolis, 138648 Singapore; dDermatology Branch, National Institute of Arthritis and Musculoskeletal and Skin Diseases, National Institutes of Health, Bethesda, MD 20892; and eDepartment of Orthopedic Surgery, National Defense Medical College, Tokorozawa 359-8513, Japan Edited by Kenneth M. Murphy, Washington University School of Medicine, St. Louis, MO, and approved June 10, 2019 (received for review November 4, 2018) Conventional dendritic cells (cDCs) derive from bone marrow (BM) intocDC1sorcDC2stakesplaceintheBM(3),andthese precursors that undergo cascades of developmental programs to pre-cDC1s and pre-cDC2s ultimately differentiate into cDC1s terminally differentiate in peripheral tissues. Pre-cDC1s and pre- and cDC2s after migrating to nonlymphoid and lymphoid tissues. + + cDC2s commit in the BM to each differentiate into CD8α /CD103 cDCs are short-lived, and their homeostatic maintenance relies + cDC1s and CD11b cDC2s, respectively. Although both cDCs rely on on constant replenishment from the BM precursors (5). The cy- the cytokine FLT3L during development, mechanisms that ensure tokine Fms-related tyrosine kinase 3 ligand (FLT3L) (12), by cDC accessibility to FLT3L have yet to be elucidated. Here, we gen- signaling through its receptor FLT3 expressed on DC precursors, erated mice that lacked a disintegrin and metalloproteinase (ADAM) is essential during the development of DCs (7, 13). -
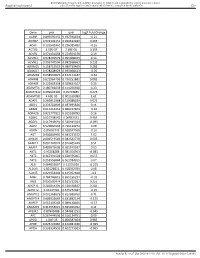
Gene Pval Qval Log2 Fold Change AAMP 0.895690332 0.952598834
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene pval qval Log2 Fold Change AAMP 0.895690332 0.952598834 -0.21 ABI3BP 0.002302151 0.020612283 0.465 ACHE 0.103542461 0.296385483 -0.16 ACTG2 2.99E-07 7.68E-05 3.195 ACVR1 0.071431098 0.224504378 0.19 ACVR1C 0.978209579 0.995008423 0.14 ACVRL1 0.006747504 0.042938663 0.235 ADAM15 0.158715519 0.380719469 0.285 ADAM17 0.978208929 0.995008423 -0.05 ADAM28 0.038932876 0.152174187 -0.62 ADAM8 0.622964796 0.790251882 0.085 ADAM9 0.122003358 0.329623107 0.25 ADAMTS1 0.180766659 0.414256926 0.23 ADAMTS12 0.009902195 0.05703885 0.425 ADAMTS8 4.60E-05 0.001169089 1.61 ADAP1 0.269811968 0.519388039 0.075 ADD1 0.233702809 0.487695826 0.11 ADM2 0.012213453 0.066227879 -0.36 ADRA2B 0.822777921 0.915518785 0.16 AEBP1 0.010738542 0.06035531 0.465 AGGF1 0.117946691 0.320915024 -0.095 AGR2 0.529860903 0.736120272 0.08 AGRN 0.85693743 0.928047568 -0.16 AGT 0.006849995 0.043233572 1.02 AHNAK 0.006519543 0.042542779 0.605 AKAP12 0.001747074 0.016405449 0.51 AKAP2 0.409929603 0.665919397 0.05 AKT1 0.95208288 0.985354963 -0.085 AKT2 0.367391504 0.620376005 0.055 AKT3 0.253556844 0.501934205 0.07 ALB 0.064833867 0.21195036 -0.315 ALDOA 0.83128831 0.918352939 0.08 ALOX5 0.029954404 0.125352668 -0.3 AMH 0.784746815 0.895196237 -0.03 ANG 0.050500474 0.181732067 0.255 ANGPT1 0.281853305 0.538528647 0.285 ANGPT2 0.43147281 0.675272487 -0.15 ANGPTL2 0.001368876 0.013688762 0.71 ANGPTL4 0.686032669 0.831882134 -0.175 ANPEP 0.019103243 0.089148466 -0.57 ANXA2P2 0.412553021 0.665966092 0.11 AP1M2 0.87843088 0.944681253 -0.045 APC 0.267444505 0.516134751 0.09 APOD 1.04E-05 0.000587404 0.985 APOE 0.023722987 0.104981036 -0.395 APOH 0.336334555 0.602273505 -0.065 Sundar R, et al. -

Epigenetic Suppression of Neprilysin Regulates Breast Cancer Invasion
OPEN Citation: Oncogenesis (2016) 5, e207; doi:10.1038/oncsis.2016.16 www.nature.com/oncsis ORIGINAL ARTICLE Epigenetic suppression of neprilysin regulates breast cancer invasion HM Stephen, RJ Khoury, PR Majmudar, T Blaylock1, K Hawkins1, MS Salama1, MD Scott1, B Cosminsky1, NK Utreja1, J Britt and RE Conway In women, invasive breast cancer is the second most common cancer and the second cause of cancer-related death. Therefore, identifying novel regulators of breast cancer invasion could lead to additional biomarkers and therapeutic targets. Neprilysin, a cell-surface enzyme that cleaves and inactivates a number of substrates including endothelin-1 (ET1), has been implicated in breast cancer, but whether neprilysin promotes or inhibits breast cancer cell progression and metastasis is unclear. Here, we asked whether neprilysin expression predicts and functionally regulates breast cancer cell invasion. RT–PCR and flow cytometry analysis of MDA-MB-231 and MCF-7 breast cancer cell lines revealed decreased neprilysin expression compared with normal epithelial cells. Expression was also suppressed in invasive ductal carcinoma (IDC) compared with normal tissue. In addition, in vitro invasion assays demonstrated that neprilysin overexpression decreased breast cancer cell invasion, whereas neprilysin suppression augmented invasion. Furthermore, inhibiting neprilysin in MCF-7 breast cancer cells increased ET1 levels significantly, whereas overexpressing neprilysin decreased extracellular-signal related kinase (ERK) activation, indicating that neprilysin negatively regulates ET1-induced activation of mitogen-activated protein kinase (MAPK) signaling. To determine whether neprilysin was epigenetically suppressed in breast cancer, we performed bisulfite conversion analysis of breast cancer cells and clinical tumor samples. We found that the neprilysin promoter was hypermethylated in breast cancer; chemical reversal of methylation in MDA-MB-231 cells reactivated neprilysin expression and inhibited cancer cell invasion. -

R&D Assay for Alzheimer's Disease
R&DR&D assayassay forfor Alzheimer’sAlzheimer’s diseasedisease Target screening⳼ Ⲽ㬔 antibody array, ᢜ⭉㬔 ⸽ἐⴐ Amyloid β-peptide Alzheimer’s disease⯸ ኸᷠ᧔ ᆹ⸽ inhibitor, antibody, ELISA kit Surwhrph#Surilohu#Dqwlerg|#Duud| 6OUSFBUFE 1."5SFBUFE )41 $3&# &3, &3, )41 $3&# &3, &3, 壤伡庰䋸TBNQMF ɅH 侴䋸嵄䍴䋸BOBMZUFT䋸䬱娴哜塵 1$ 1$ 1$ 1$ 5IFNPTUSFGFSFODFEBSSBZT 1$ 1$ QQ α 34, .4, 503 Q α 34, .4, 503 %SVHTDSFFOJOH0òUBSHFUFòFDUT0ATHWAY涭廐 6OUSFBUFE 堄币䋸4BNQMF侴䋸8FTUFSOPS&-*4"䍘䧽 1."5SFBUFE P 8FTUFSOCMPU廽喜儤应侴䋸0, Z 4VCTUSBUF -JHIU )31DPOKVHBUFE1BO "OUJQIPTQIPUZSPTJOF .FBO1JYFM%FOTJUZ Y $BQUVSF"OUJCPEZ 5BSHFU"OBMZUF "SSBZ.FNCSBOF $3&# &3, &3, )41 .4, Q α 34, 503 Human XL Cytokine Array kit (ARY022, 102 analytes) Adiponectin,Aggrecan,Angiogenin,Angiopoietin-1,Angiopoietin-2,BAFF,BDNF,Complement,Component C5/C5a,CD14,CD30,CD40L, Chitinase 3-like 1,Complement Factor D,C-Reactive Protein,Cripto-1,Cystatin C,Dkk-1,DPPIV,EGF,EMMPRIN,ENA-78,Endoglin, Fas L,FGF basic,FGF- 7,FGF-19,Flt-3 L,G-CSF,GDF-15,GM-CSF,GRO-α,Grow th Hormone,HGF,ICAM-1,IFN-γ,IGFBP-2,IGFBP-3, IL-1α,IL-1β, IL-1ra,IL-2,IL-3,IL-4,IL- 5,IL-6,IL-8, IL-10,IL-11,IL-12, IL-13,IL-15,IL-16,IL-17A,IL-18 BPa,IL-19,IL-22, IL-23,IL-24,IL-27, IL-31,IL-32α/β/γ,IL-33,IL-34,IP-10,I-TAC,Kallikrein 3,Leptin,LIF,Lipocalin-2,MCP-1,MCP-3,M-CSF,MIF,MIG,MIP-1α/MIP-1β,MIP-3α,MIP-3β,MMP-9, Myeloperoxidase,Osteopontin, p70, PDGF-AA, PDGF-AB/BB,Pentraxin-3, PF4, RAGE, RANTES,RBP4,Relaxin-2, Resistin,SDF-1α,Serpin E1, SHBG, ST2, TARC,TFF3,TfR,TGF- ,Thrombospondin-1,TNF-α, uPAR, VEGF, Vitamin D BP Human Protease (34 analytes) / -

Gene Standard Deviation MTOR 0.12553731 PRPF38A
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut Gene Standard Deviation MTOR 0.12553731 PRPF38A 0.141472605 EIF2B4 0.154700091 DDX50 0.156333027 SMC3 0.161420017 NFAT5 0.166316903 MAP2K1 0.166585267 KDM1A 0.16904912 RPS6KB1 0.170330192 FCF1 0.170391706 MAP3K7 0.170660513 EIF4E2 0.171572093 TCEB1 0.175363093 CNOT10 0.178975095 SMAD1 0.179164705 NAA15 0.179904998 SETD2 0.180182498 HDAC3 0.183971158 AMMECR1L 0.184195031 CHD4 0.186678211 SF3A3 0.186697697 CNOT4 0.189434633 MTMR14 0.189734199 SMAD4 0.192451524 TLK2 0.192702667 DLG1 0.19336621 COG7 0.193422331 SP1 0.194364189 PPP3R1 0.196430217 ERBB2IP 0.201473001 RAF1 0.206887192 CUL1 0.207514271 VEZF1 0.207579584 SMAD3 0.208159809 TFDP1 0.208834504 VAV2 0.210269344 ADAM17 0.210687138 SMURF2 0.211437666 MRPS5 0.212428684 TMUB2 0.212560675 SRPK2 0.216217428 MAP2K4 0.216345366 VHL 0.219735582 SMURF1 0.221242495 PLCG1 0.221688351 EP300 0.221792349 Sundar R, et al. Gut 2020;0:1–10. doi: 10.1136/gutjnl-2020-320805 BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) Gut MGAT5 0.222050228 CDC42 0.2230598 DICER1 0.225358787 RBX1 0.228272533 ZFYVE16 0.22831803 PTEN 0.228595789 PDCD10 0.228799406 NF2 0.23091035 TP53 0.232683696 RB1 0.232729172 TCF20 0.2346075 PPP2CB 0.235117302 AGK 0.235416298