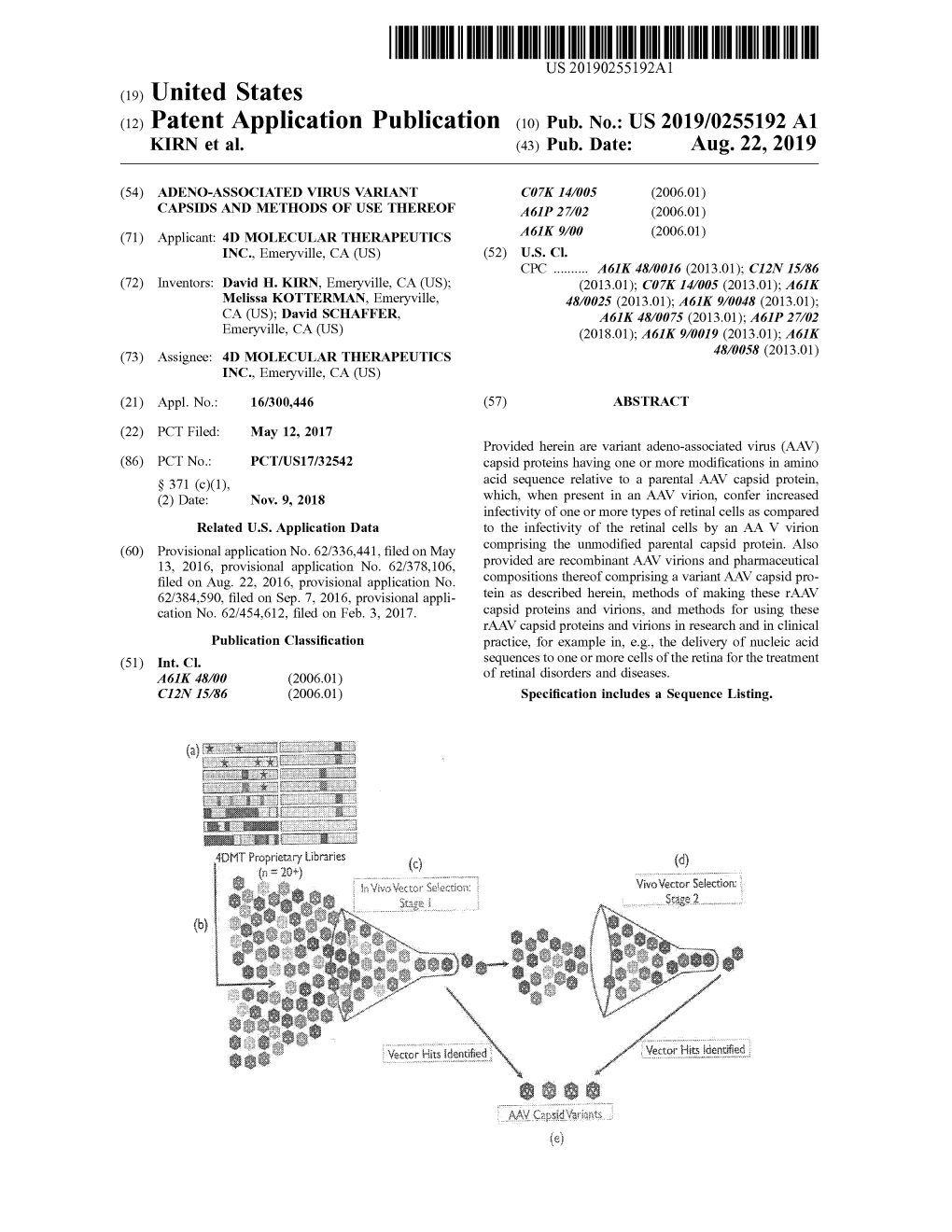
Transmitimijeni Vector Kits Identified Vector???? Hits ?? ???? ???? ???? ?? ?? ?? ?????Identified ??? : : ??? ?? ? 900000 I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Organ Level Protein Networks As a Reference for the Host Effects of the Microbiome
Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 1 Organ level protein networks as a reference for the host effects of the microbiome 2 3 Robert H. Millsa,b,c,d, Jacob M. Wozniaka,b, Alison Vrbanacc, Anaamika Campeaua,b, Benoit 4 Chassainge,f,g,h, Andrew Gewirtze, Rob Knightc,d, and David J. Gonzaleza,b,d,# 5 6 a Department of Pharmacology, University of California, San Diego, California, USA 7 b Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California, San Diego, 8 California, USA 9 c Department of Pediatrics, and Department of Computer Science and Engineering, University of 10 California, San Diego California, USA 11 d Center for Microbiome Innovation, University of California, San Diego, California, USA 12 e Center for Inflammation, Immunity and Infection, Institute for Biomedical Sciences, Georgia State 13 University, Atlanta, GA, USA 14 f Neuroscience Institute, Georgia State University, Atlanta, GA, USA 15 g INSERM, U1016, Paris, France. 16 h Université de Paris, Paris, France. 17 18 Key words: Microbiota, Tandem Mass Tags, Organ Proteomics, Gnotobiotic Mice, Germ-free Mice, 19 Protein Networks, Proteomics 20 21 # Address Correspondence to: 22 David J. Gonzalez, PhD 23 Department of Pharmacology and Pharmacy 24 University of California, San Diego 25 La Jolla, CA 92093 26 E-mail: [email protected] 27 Phone: 858-822-1218 28 1 Downloaded from genome.cshlp.org on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press 29 Abstract 30 Connections between the microbiome and health are rapidly emerging in a wide range of 31 diseases. -

Miasdb: a Database of Molecular Interactions Associated with Alternative Splicing of Human Pre-Mrnas
RESEARCH ARTICLE MiasDB: A Database of Molecular Interactions Associated with Alternative Splicing of Human Pre-mRNAs Yongqiang Xing1, Xiujuan Zhao1, Tao Yu2, Dong Liang1, Jun Li1, Guanyun Wei1, Guoqing Liu1, Xiangjun Cui1, Hongyu Zhao1, Lu Cai1* 1 School of Life Science and Technology, Inner Mongolia University of Science and Technology, Baotou, 014010, China, 2 School of Science, Inner Mongolia University of Science and Technology, Baotou, 014010, China a11111 * [email protected] Abstract Alternative splicing (AS) is pervasive in human multi-exon genes and is a major contributor to expansion of the transcriptome and proteome diversity. The accurate recognition of alter- OPEN ACCESS native splice sites is regulated by information contained in networks of protein-protein and Citation: Xing Y, Zhao X, Yu T, Liang D, Li J, Wei G, protein-RNA interactions. However, the mechanisms leading to splice site selection are not et al. (2016) MiasDB: A Database of Molecular fully understood. Although numerous databases have been built to describe AS, molecular Interactions Associated with Alternative Splicing of Human Pre-mRNAs. PLoS ONE 11(5): e0155443. interaction databases associated with AS have only recently emerged. In this study, we doi:10.1371/journal.pone.0155443 present a new database, MiasDB, that provides a description of molecular interactions Editor: Ruben Artero, University of Valencia, SPAIN associated with human AS events. This database covers 938 interactions between human splicing factors, RNA elements, transcription factors, kinases and modified histones for 173 Received: November 19, 2015 human AS events. Every entry includes the interaction partners, interaction type, experi- Accepted: April 28, 2016 mental methods, AS type, tissue specificity or disease-relevant information, a simple Published: May 11, 2016 description of the functionally tested interaction in the AS event and references. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -
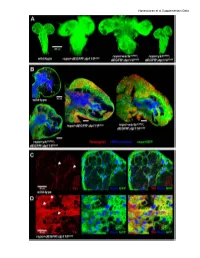
Supplementary Data Vigneswaran Et Al Supplementary Data
Vigneswaran et al Supplementary Data Vigneswaran et al Supplementary Data Figure S1: Yki is required for EGFR-PI3K-driven glial neoplasia in Drosophila (A) Optical projections of whole brain-nerve cord complexes from 3rd instar larvae approximately 130 hrs old. Dorsal view; anterior up. CD8-GFP (green) labels glial cell bodies. Compared to repo>dEGFRλ;dp110CAAX, warts knockdown (repo>wartsdsRNA; dEGFRλ;dp110CAAX) increased neoplastic brain overgrowth and yki knockdown (repo>ykidsRNA;dEGFRλ;dp110CAAX) decreased neoplastic brain overgrowth. (B) 3 µm optical projections of brain hemispheres, age-matched 3rd instar larvae. Frontal sections; anterior up; midline to left. Repo (red) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies; anti-HRP (blue) counter-stains for neurons and neuropil. (middle) repo>dEGFRλ;dp110CAAX showed increased glial cell numbers (red nuclei) compared to (upper left) wild-type. Compared to repo>dEGFRλ;dp110CAAX, (right) warts knockdown increased neoplastic glial cell numbers (red nuclei), whereas (lower left) yki knockdown reduced neoplastic glial cell numbers (red nuclei). (C, D) Low levels of Yki protein (red) was observed in wild-type central brain glia (white arrows, left panel in C) compared to high levels of cytoplasmic and nuclear Yki protein in dEGFRλ;dp110CAAX neoplastic glia (white arrows, left panel in D); Repo (blue) labels glial cell nuclei; CD8-GFP (green) labels glial cell bodies. Vigneswaran et al Supplementary Data Figure S2: YAP/TAZ expression confined to RTK-amplified tuMor cells and Maintained in patient-derived xenografts (A) On the left, immunohistochemical (IHC) staining in representative normal brain parenchyma in the cortex where YAP expression and TAZ expression was limited to vascular cells and was not detectable in normal neuronal and glial cells. -

Targeted Exome Capture and Sequencing Identifies Novel
Open Access Research BMJ Open: first published as 10.1136/bmjopen-2013-004030 on 7 November 2013. Downloaded from Targeted exome capture and sequencing identifies novel PRPF31 mutations in autosomal dominant retinitis pigmentosa in Chinese families Liping Yang,1 Xiaobei Yin,2 Lemeng Wu,1 Ningning Chen,1 Huirong Zhang,1 Genlin Li,2 Zhizhong Ma1 To cite: Yang L, Yin X, Wu L, ABSTRACT et al Strengths and limitations of this study . Targeted exome capture Objectives: To identify disease-causing mutations in and sequencing identifies two Chinese families with autosomal dominant retinitis ▪ The HEDEP based on targeted exome capture novel PRPF31 mutations in pigmentosa (adRP). autosomal dominant retinitis technology is an efficient method for molecular pigmentosa in Chinese Design: Prospective analysis. diagnosis in adRP patients. families. BMJ Open 2013;3: Patients: Two Chinese adRP families underwent ▪ Both mutations result in premature termination e004030. doi:10.1136/ genetic diagnosis. A specific hereditary eye disease codons before the last exon, thus insufficient bmjopen-2013-004030 enrichment panel (HEDEP) based on targeted exome functioning due to haploinsufficiency instead of capture technology was used to collect the protein aberrant function of the mutated proteins seems ▸ Prepublication history and coding regions of targeted 371 hereditary eye disease to be the most probably reason in these two additional material for this genes; high throughput sequencing was done with the families. However no experiment was done to paper is available online. To Illumina HiSeq 2000 platform. The identified variants prove it in this study. view these files please visit were confirmed with Sanger sequencing. the journal online Setting: All experiments were performed in a large (http://dx.doi.org/10.1136/ laboratory specialising in genetic studies in the bmjopen-2013-004030). -

Host Cell Factors Necessary for Influenza a Infection: Meta-Analysis of Genome Wide Studies
Host Cell Factors Necessary for Influenza A Infection: Meta-Analysis of Genome Wide Studies Juliana S. Capitanio and Richard W. Wozniak Department of Cell Biology, Faculty of Medicine and Dentistry, University of Alberta Abstract: The Influenza A virus belongs to the Orthomyxoviridae family. Influenza virus infection occurs yearly in all countries of the world. It usually kills between 250,000 and 500,000 people and causes severe illness in millions more. Over the last century alone we have seen 3 global influenza pandemics. The great human and financial cost of this disease has made it the second most studied virus today, behind HIV. Recently, several genome-wide RNA interference studies have focused on identifying host molecules that participate in Influen- za infection. We used nine of these studies for this meta-analysis. Even though the overlap among genes identified in multiple screens was small, network analysis indicates that similar protein complexes and biological functions of the host were present. As a result, several host gene complexes important for the Influenza virus life cycle were identified. The biological function and the relevance of each identified protein complex in the Influenza virus life cycle is further detailed in this paper. Background and PA bound to the viral genome via nucleoprotein (NP). The viral core is enveloped by a lipid membrane derived from Influenza virus the host cell. The viral protein M1 underlies the membrane and anchors NEP/NS2. Hemagglutinin (HA), neuraminidase Viruses are the simplest life form on earth. They parasite host (NA), and M2 proteins are inserted into the envelope, facing organisms and subvert the host cellular machinery for differ- the viral exterior. -

The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease
International Journal of Molecular Sciences Review The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease Izarbe Aísa-Marín 1,2 , Rocío García-Arroyo 1,3 , Serena Mirra 1,2 and Gemma Marfany 1,2,3,* 1 Departament of Genetics, Microbiology and Statistics, Avda. Diagonal 643, Universitat de Barcelona, 08028 Barcelona, Spain; [email protected] (I.A.-M.); [email protected] (R.G.-A.); [email protected] (S.M.) 2 Centro de Investigación Biomédica en Red Enfermedades Raras (CIBERER), Instituto de Salud Carlos III (ISCIII), Universitat de Barcelona, 08028 Barcelona, Spain 3 Institute of Biomedicine (IBUB, IBUB-IRSJD), Universitat de Barcelona, 08028 Barcelona, Spain * Correspondence: [email protected] Abstract: Alternative splicing of mRNA is an essential mechanism to regulate and increase the diversity of the transcriptome and proteome. Alternative splicing frequently occurs in a tissue- or time-specific manner, contributing to differential gene expression between cell types during development. Neural tissues present extremely complex splicing programs and display the highest number of alternative splicing events. As an extension of the central nervous system, the retina constitutes an excellent system to illustrate the high diversity of neural transcripts. The retina expresses retinal specific splicing factors and produces a large number of alternative transcripts, including exclusive tissue-specific exons, which require an exquisite regulation. In fact, a current challenge in the genetic diagnosis of inherited retinal diseases stems from the lack of information regarding alternative splicing of retinal genes, as a considerable percentage of mutations alter splicing Citation: Aísa-Marín, I.; or the relative production of alternative transcripts. Modulation of alternative splicing in the retina García-Arroyo, R.; Mirra, S.; Marfany, is also instrumental in the design of novel therapeutic approaches for retinal dystrophies, since it G. -

Mrna Editing, Processing and Quality Control in Caenorhabditis Elegans
| WORMBOOK mRNA Editing, Processing and Quality Control in Caenorhabditis elegans Joshua A. Arribere,*,1 Hidehito Kuroyanagi,†,1 and Heather A. Hundley‡,1 *Department of MCD Biology, UC Santa Cruz, California 95064, †Laboratory of Gene Expression, Medical Research Institute, Tokyo Medical and Dental University, Tokyo 113-8510, Japan, and ‡Medical Sciences Program, Indiana University School of Medicine-Bloomington, Indiana 47405 ABSTRACT While DNA serves as the blueprint of life, the distinct functions of each cell are determined by the dynamic expression of genes from the static genome. The amount and specific sequences of RNAs expressed in a given cell involves a number of regulated processes including RNA synthesis (transcription), processing, splicing, modification, polyadenylation, stability, translation, and degradation. As errors during mRNA production can create gene products that are deleterious to the organism, quality control mechanisms exist to survey and remove errors in mRNA expression and processing. Here, we will provide an overview of mRNA processing and quality control mechanisms that occur in Caenorhabditis elegans, with a focus on those that occur on protein-coding genes after transcription initiation. In addition, we will describe the genetic and technical approaches that have allowed studies in C. elegans to reveal important mechanistic insight into these processes. KEYWORDS Caenorhabditis elegans; splicing; RNA editing; RNA modification; polyadenylation; quality control; WormBook TABLE OF CONTENTS Abstract 531 RNA Editing and Modification 533 Adenosine-to-inosine RNA editing 533 The C. elegans A-to-I editing machinery 534 RNA editing in space and time 535 ADARs regulate the levels and fates of endogenous dsRNA 537 Are other modifications present in C. -

Disrupted Alternative Splicing for Genes Implicated in Splicing and Ciliogenesis Causes PRPF31 Retinitis Pigmentosa
ARTICLE DOI: 10.1038/s41467-018-06448-y OPEN Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa Adriana Buskin et al.# Mutations in pre-mRNA processing factors (PRPFs) cause autosomal-dominant retinitis pigmentosa (RP), but it is unclear why mutations in ubiquitously expressed genes cause non- 1234567890():,; syndromic retinal disease. Here, we generate transcriptome profiles from RP11 (PRPF31- mutated) patient-derived retinal organoids and retinal pigment epithelium (RPE), as well as Prpf31+/− mouse tissues, which revealed that disrupted alternative splicing occurred for specific splicing programmes. Mis-splicing of genes encoding pre-mRNA splicing proteins was limited to patient-specific retinal cells and Prpf31+/− mouse retinae and RPE. Mis-splicing of genes implicated in ciliogenesis and cellular adhesion was associated with severe RPE defects that include disrupted apical – basal polarity, reduced trans-epithelial resistance and phagocytic capacity, and decreased cilia length and incidence. Disrupted cilia morphology also occurred in patient-derived photoreceptors, associated with progressive degeneration and cellular stress. In situ gene editing of a pathogenic mutation rescued protein expression and key cellular phenotypes in RPE and photoreceptors, providing proof of concept for future therapeutic strategies. Correspondence and requests for materials should be addressed to S.-N.G. (email: [email protected]) or to C.A.J. (email: [email protected]) or to M.L. (email: [email protected]). #A full list of authors and their affliations appears at the end of the paper. NATURE COMMUNICATIONS | (2018) 9:4234 | DOI: 10.1038/s41467-018-06448-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | DOI: 10.1038/s41467-018-06448-y etinitis pigmentosa (RP) is one of the most common c.522_527+10del heterozygous mutation (Supplementary Rinherited forms of retinal blindness with a prevalence of Data 1). -

RBMX Family Proteins Connect the Fields of Nuclear RNA Processing
International Journal of Biochemistry and Cell Biology 108 (2019) 1–6 Contents lists available at ScienceDirect International Journal of Biochemistry and Cell Biology journal homepage: www.elsevier.com/locate/biocel RBMX family proteins connect the fields of nuclear RNA processing, disease and sex chromosome biology T ⁎ David J. Elliott , Caroline Dalgliesh, Gerald Hysenaj, Ingrid Ehrmann Institute of Genetic Medicine, Newcastle University, Newcastle-upon-Tyne, NE1 3BZ, UK ARTICLE INFO ABSTRACT Keywords: RBMX is a ubiquitously expressed nuclear RNA binding protein that is encoded by a gene on the X chromosome. RNA splicing RBMX belongs to a small protein family with additional members encoded by paralogs on the mammalian Y Gene expression chromosome and other chromosomes. These RNA binding proteins are important for normal development, and Brain development also implicated in cancer and viral infection. At the molecular level RBMX family proteins contribute to splicing Testis control, transcription and genome integrity. Establishing what endogenous genes and pathways are controlled by Cancer RBMX and its paralogs will have important implications for understanding chromosome biology, DNA repair and mammalian development. Here we review what is known about this family of RNA binding proteins, and identify important current questions about their functions. 1. Introduction et al., 1993). While RBMX is expressed ubiquitously through the body, RBMY genes are specifically expressed in germ cells (cells that even- RBMX (an acronym of RNA Binding Motif protein, X-linked) was tually develop into sperm). Multiple RBMY genes are present on the originally identified as one of a functionally diverse group of nuclear long arm of the human Y chromosome, each encoding 496 amino acid proteins that bind to polyadenylated RNA. -

Supplemental Table 2
Supplemental Table 2 probeID geneSymbol geneTitle cox.tt 223518_at DFFA DNA fragmentation factor, 45kDa, alpha polypeptide 4.86807974 212370_x_at FAM21A /// FAM21B /// FAM21C family with sequence similarity 21, member A /// family with sequence similarity 21, member B /// family with sequence similarity 21, member C -4.039911936 228937_at C13orf31 chromosome 13 open reading frame 31 -3.965652752 224217_s_at FAF1 Fas (TNFRSF6) associated factor 1 3.818692986 219066_at PPCDC phosphopantothenoylcysteine decarboxylase 3.812463041 200994_at IPO7 importin 7 3.722967297 203853_s_at GAB2 GRB2-associated binding protein 2 -3.576924648 218118_s_at TIMM23 translocase of inner mitochondrial membrane 23 homolog (yeast) -3.525051556 238662_at ATPBD4 ATP binding domain 4 3.522716621 212905_at CSTF2T cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant -3.458703211 225075_at PDRG1 p53 and DNA-damage regulated 1 3.437886045 211430_s_at IGH@ /// IGHG1 /// IGHG2 /// IGHM /// IGHV4-31 /// LOC100294459 immunoglobulin heavy locus /// immunoglobulin heavy constant gamma 1 (G1m marker) /// immunoglobulin heavy constant gamma 2 (G2m marker) /// immunoglobulin heavy constant mu /// immunoglobulin heavy variable 4-31 /// similar to immunoglobulin lambda-like polypeptide 1 -3.404297079 244181_at 244181_at --- -3.363118329 202231_at EIF3M eukaryotic translation initiation factor 3, subunit M 3.345657723 201160_s_at CSDA cold shock domain protein A 3.337512802 214946_x_at FAM21A /// FAM21B /// FAM21C /// FAM21D family with sequence similarity 21,