Report on Existing Open-Source Electronic Medical Records
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

Examples of the Electronic Health Record (EHR)
Examples of the Electronic Health Record (EHR) What is Biomedical & Health Informatics? William Hersh Copyright 2020 Oregon Health & Science University Examples of the EHR • Using the Veterans Health Information Systems and Technology Architecture (VistA) • Why VistA? – State-of-the-art EHR that has transformed healthcare in the Veteran’s Health Administration (VHA) (Perlin, 2006; Byrne, 2010) – Not that pretty, but has all of the modern features of the EHR, e.g., clinical decision support (CDS), computerized provider order entry (CPOE), etc. – Distributed under open-source model, unlike most other vendors who do not even allow screen shots to be shown outside their customers’ institutions • Fascinating story (Allen, 2017), but VistA now being phased out in favor of Cerner EHR adopted by Department of Defense (VA, 2017) – https://www.ehrm.va.gov/ WhatIs5 2 1 Some details about VistA • Server written in M (formerly called MUMPS), accessed via command-line interface – Runs in commercial Intersystems Cache (on many platforms) or open-source GT.M (Linux only) • Client (called CPRS) written in Delphi and providers graphical user interface – Only runs on Windows (just about all versions) WhatIs5 3 Logging on to CPRS, the front end to VistA WhatIs5 4 2 Choosing a patient WhatIs5 5 Cover sheet – overview WhatIs5 6 3 Drilling down to details of a problem WhatIs5 7 Details of an allergy WhatIs5 8 4 Viewing vital signs over time WhatIs5 9 More details on problems WhatIs5 10 5 List of active orders WhatIs5 11 Viewing the patient’s notes WhatIs5 12 -

Open Source As an Alternative for Clinical Information Systems Adoption
Open Source as an Alternative for Clinical Information Systems Adoption Placide Poba-Nzaou , Université du Québec à Montréal Josianne Marsan , Université Laval Guy Paré , HEC Montréal Louis Raymond , Université du Québec à Trois-Rivières Health Informatics & Technology Conference October 20-22, 2014 - Baltimore, MD, USA Agenda • Context • Research Questions • Conceptual Background • Research Method • Results • Discussion • Conclusion 2 Context • Electronic Health Records (EHR) are at the heart of most health system reforms – “Mission-critical” applications for hospitals • High costs and low level of interoperability of commercial EHR software has led a growing number of hospitals to adopt open source software (OSS) solutions – Examples of OSS EHR: VistA, Oscar, GNU Med, OpenEMR and OpenMRS, etc. 3 Research Questions • What can motivate a Hospital to adopt an Open Source EHR ? • What are the main challenges faced by a hospital adopting an Open Source EHR ? • What can be done to deal with these challenges ? 4 Conceptual Background • Motivations to adopt an Enterprise System by healthcare organizations (Poba-Nzaou et al., 2014) – Strategic (Clinical vs. Managerial) – Operational (Clinical vs. Managerial) – Financial – Technological • Hybrid organization – “Organization that combine different institutional logics in unprecedented ways” (Scott and Meyer 1991) • E.g.: integration of not-for-profit and for-profit logics (Battilana et al., 2012) – Challenges faced by Hybrid Organizations in search of sustainability (Battilana et al., 2012) – #1: Legal Structure – #2: Financing – #3: Customers and Beneficiaries 5 – #4: Organizational Culture and Talent Development Conceptual Background (continued) – OSS governance models • “the means of achieving the direction, control, and coordination of wholly or partially autonomous individuals and organizations on behalf of an OSS development project to which they jointly contribute” (Markus, 2007) – “community managed” vs. -

Mobile Applications for the Health Sector
Mobile Applications for the Health Sector Christine Zhenwei Qiang, Masatake Yamamichi*, Vicky Hausman and Daniel Altman ICT Sector Unit World Bank December 2011 This report is the product of the staff and consultants of the World Bank. The findings, interpretations, and conclusions do not necessarily reflect the views of the Executive Directors of the World Bank or the governments they represent. The World Bank does not guarantee the accuracy of the data included in this work. * Corresponding author: 1818 H Street NW, MSN MC6-616, Washington DC 20433, USA. [email protected] Contents Abbreviations ................................................................................................................... 3 Acknowledgements ........................................................................................................... 4 Executive Summary ........................................................................................................... 5 1 Introduction ........................................................................................................... 11 1.1 What is mobile health? 11 1.2 Technological context for mobile health 12 1.3 Perceived potential of mobile health 13 1.4 The mobile health ecosystem 16 1.5 Social goals of investments in mobile health 17 1.6 How does mobile health relate to other intersections of health and technology? 19 2 Health Needs in Developing Countries ...................................................................... 21 2.1 Common health burdens 21 2.2 Challenges -

A Cloud Based Patient-Centered Ehealth Record
International Journal on Advances in Life Sciences, vol 7 no 1 & 2, year 2015, http://www.iariajournals.org/life_sciences/ 30 A Cloud Based Patient-Centered eHealth Record Konstantinos Koumaditis, Leonidas Katelaris, and Marinos Themistocleous Department of Digital Systems University of Piraeus Piraeus, Greece e-mail: (konkoum,lkatelaris,mthemist)@unipi.gr Abstract - This research focuses on the Patient-Centered integrated view of citizens health status. The latter is reflected e-Health (PCEH) concept by introducing its importance in EHRs and Personal Health Records (PHRs), which are and demonstrating a multidisciplinary project that being enriched and exploited by different actors and combines advanced technologies. The project links stakeholders (i.e., health and care professionals, citizens, several aspects of PCEH functionality, such as: (a) nutrition experts, hospitals, etc.) in the health ecosystem. homecare telemedicine technologies, (b) e-prescribing, e- Three general PHR models have been proposed [7]: a) the referral, e-learning and (c) state-of-the-art technologies stand-alone model, b) Electronic Health Record (EHR) like cloud computing and Service Oriented Architecture system, and c) the integrated one, which is an interoperable (SOA), will lead to an innovative integrated e-health system providing linkage with a variety of patient platform that delivers many benefits to the society, the information sources, such as EHRs, home diagnostics, economy, the industry and the research community. This insurance claims etc. The main types of health information paper provides insights of the PCEH concept and the supported by PHRs are problem lists, procedures, major current stages of the project. In doing so, we aim to illnesses, provider lists, allergy data, home-monitored data, increase the awareness of this significant work and family history, social history and lifestyle, immunizations, disseminate the knowledge gained so far through our medications and laboratory tests [8, 9]. -

Debian Med Integrated Software Environment for All Medical Applications
Debian Med Integrated software environment for all medical applications Andreas Tille 27. February 2013 When people hear for the first time the term ‘Debian Med’ there are usually two kinds of misconceptions. Let us dispel these in advance, so as to clarify subsequent discussion of the project. People familiar with Debian as a large distribution of Free Software usually imag- ine Debian Med to be some kind of customised derivative of Debian tailored for use in a medical environment. Astonishingly, the idea that such customisation can be done entirely within Debian itself is not well known and the technical term Debian Pure Blend seems to be sufficiently unknown outside of the Debian milieu that many people fail to appreciate the concept correctly. There are no separate repositories like Personal Package Archives (PPA) as introduced by Ubuntu for additional software not belong- ing to the official distribution or something like that – a Debian Pure Blend (as the term ’pure’ implies) is Debian itself and if you have received Debian you have full De- bian Med at your disposal. There are other Blends inside Debian like Debian Science, Debian Edu, Debian GIS and others. People working in the health care professions sometimes acquire another miscon- ception about Debian Med, namely that Debian Med is some kind of software primarily dedicated to managing a doctor’s practice. Sometimes people even assume that people assume the Debian Med team actually develops this software. However, the truth about the Debian Med team is that we are a group of Debian developers hard at work incor- porating existing medical software right into the Debian distribution. -

Linux: Come E Perchх
ÄÒÙÜ Ô ©2007 mcz 12 luglio 2008 ½º I 1. Indice II ½º Á ¾º ¿º ÈÖÞÓÒ ½ º È ÄÒÙÜ ¿ º ÔÔÖÓÓÒÑÒØÓ º ÖÒÞ ×Ó×ØÒÞÐ ÏÒÓÛ× ¾½ º ÄÒÙÜ ÕÙÐ ×ØÖÙÞÓÒ ¾ º ÄÒÙÜ ÀÖÛÖ ×ÙÔÔ ÓÖØØÓ ¾ º È Ð ÖÒÞ ØÖ ÖÓ ÓØ Ù×Ö ¿½ ½¼º ÄÒÙÜ × Ò×ØÐÐ ¿¿ ½½º ÓÑ × Ò×ØÐÐÒÓ ÔÖÓÖÑÑ ¿ ½¾º ÒÓÒ ØÖÓÚÓ ÒÐ ×ØÓ ÐÐ ×ØÖÙÞÓÒ ¿ ½¿º Ó׳ ÙÒÓ ¿ ½º ÓÑ × Ð ××ØÑ ½º ÓÑ Ð ½º Ð× Ñ ½º Ð Ñ ØÐ ¿ ½º ÐÓ ½º ÓÑ × Ò×ØÐÐ Ð ×ØÑÔÒØ ¾¼º ÓÑ ÐØØÖ¸ Ø×Ø ÐÖ III Indice ¾½º ÓÑ ÚÖ Ð ØÐÚ×ÓÒ ¿ 21.1. Televisioneanalogica . 63 21.2. Televisione digitale (terrestre o satellitare) . ....... 64 ¾¾º ÐÑØ ¾¿º Ä 23.1. Fotoritocco ............................. 67 23.2. Grafica3D.............................. 67 23.3. Disegnovettoriale-CAD . 69 23.4.Filtricoloreecalibrazionecolori . .. 69 ¾º ×ÖÚ Ð ½ 24.1.Vari.................................. 72 24.2. Navigazionedirectoriesefiles . 73 24.3. CopiaCD .............................. 74 24.4. Editaretesto............................. 74 24.5.RPM ................................. 75 ¾º ×ÑÔ Ô ´ËÐе 25.1.Montareundiscoounapenna . 77 25.2. Trovareunfilenelsistema . 79 25.3.Vedereilcontenutodiunfile . 79 25.4.Alias ................................. 80 ¾º × ÚÓÐ×× ÔÖÓÖÑÑÖ ½ ¾º ÖÓÛ×Ö¸ ÑÐ ººº ¿ ¾º ÖÛÐРгÒØÚÖÙ× Ð ÑØØÑÓ ¾º ÄÒÙÜ ½ ¿¼º ÓÑ ØÖÓÚÖ ÙØÓ ÖÖÑÒØ ¿ ¿½º Ð Ø×ØÙÐ Ô Ö Ð ×ØÓÔ ÄÒÙÜ ¿¾º ´ÃµÍÙÒØÙ¸ ÙÒ ×ØÖÙÞÓÒ ÑÓÐØÓ ÑØ ¿¿º ËÙÜ ÙÒ³ÓØØÑ ×ØÖÙÞÓÒ ÄÒÙÜ ½¼½ ¿º Á Ó Ò ÄÒÙÜ ½¼ ¿º ÃÓÒÕÙÖÓÖ¸ ÕÙ×ØÓ ½¼ ¿º ÃÓÒÕÙÖÓÖ¸ Ñ ØÒØÓ Ô Ö ½½¿ 36.1.Unaprimaocchiata . .114 36.2.ImenudiKonqueror . .115 36.3.Configurazione . .116 IV Indice 36.4.Alcuniesempidiviste . 116 36.5.Iservizidimenu(ServiceMenu) . 119 ¿º ÃÓÒÕÙÖÓÖ Ø ½¾¿ ¿º à ÙÒ ÖÖÒØ ½¾ ¿º à ÙÒ ÐÙ×ÓÒ ½¿½ ¼º ÓÒÖÓÒØÓ Ò×ØÐÐÞÓÒ ÏÒÓÛ×È ÃÍÙÒØÙ º½¼ ½¿¿ 40.1. -

The Open Organization Guide for Educators
The Open Organization Guide for Educators The Open Organization Guide for Educators Transformative teaching and learning for a rapidly changing world Contributing authors (in alphabetical order) Amarachi Achonu Mukkai Krishnamoorthy Beth Anderson Gina Likins Bryan Behrenshausen Clara May Maxwell Bushong Ian McDermott Carolyn Butler Dan McGuire Curtis A. Carver Christopher McHale Aria F. Chernik Race MoChridhe Susie Choi Steven Ovadia Dipankar Dasgupta Ben Owens Heidi Ellis David Preston Denise Ferebee Rahul Razdan David Goldschmidt Charlie Reisinger Adam Haigler Justin Sherman Jim Hall Wesley Turner Tanner Johnson Shobha Tyagi Marcus Kelly Jim Whitehurst Ryan Williams Copyright Copyright © 2019 Red Hat, Inc. All written content, as well as the cover image, licensed under a Creative Commons Attribu- tion-ShareAlike 4.0 International License.1 Amarachi Achonu's "Making computer science curricula as adaptable as our code" originally appeared at https://opensource.- com/open-organization/19/4/adaptable-curricula-computer-science Beth Anderson's "An open process for discovering our school's core values" originally appeared at https://opensource.- com/open-organization/16/6/opening-discover-education-centers- core-values. Curtis A. Carver's "Crowdsourcing our way to a campus IT plan" originally appeared at https://opensource.com/open-organiza- tion/17/10/uab-100-wins-through-crowdsourcing. Brandon Dixon and Randall Joyce's "The most valuable cy- bersecurity education is an open one" originally appeared at https://opensource.com/open-organization/19/8/open-cybersecu- rity-education. Heidi Ellis' "What happened when I let my students fork the syllabus" originally appeared at https://opensource.com/open-orga- nization/18/11/making-course-syllabus-open. -
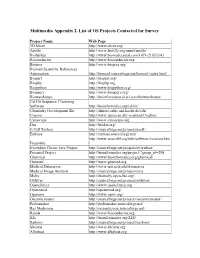
PDF File, 212 KB
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

Comparison of Open-Source Electronic Health Record Systems Based on Functional and User Performance Criteria
Original Article Healthc Inform Res. 2019 April;25(2):89-98. https://doi.org/10.4258/hir.2019.25.2.89 pISSN 2093-3681 • eISSN 2093-369X Comparison of Open-Source Electronic Health Record Systems Based on Functional and User Performance Criteria Saptarshi Purkayastha1, Roshini Allam1, Pallavi Maity1, Judy W. Gichoya2 1Department of BioHealth Informatics, Indiana University–Purdue University Indianapolis, Indianapolis, IN, USA 2Dotter Institute, Oregon Health and Science University, Portland, OR, USA Objectives: Open-source Electronic Health Record (EHR) systems have gained importance. The main aim of our research is to guide organizational choice by comparing the features, functionality, and user-facing system performance of the five most popular open-source EHR systems. Methods: We performed qualitative content analysis with a directed approach on recently published literature (2012–2017) to develop an integrated set of criteria to compare the EHR systems. The functional criteria are an integration of the literature, meaningful use criteria, and the Institute of Medicine’s functional requirements of EHR, whereas the user-facing system performance is based on the time required to perform basic tasks within the EHR system. Results: Based on the Alexa web ranking and Google Trends, the five most popular EHR systems at the time of our study were OSHERA VistA, GNU Health, the Open Medical Record System (OpenMRS), Open Electronic Medical Record (OpenEMR), and OpenEHR. We also found the trends in popularity of the EHR systems and the locations where they were more popular than others. OpenEMR met all the 32 functional criteria, OSHERA VistA met 28, OpenMRS met 12 fully and 11 partially, OpenEHR-based EHR met 10 fully and 3 partially, and GNU Health met the least with only 10 criteria fully and 2 partially. -
Step-By-Step Install Guide OSCAR Mcmaster CMS on Linux Ubuntu 10.04 LTS (Lucid Lynx)
Global Open Versity eHealth Labs Install Guide OSCAR McMaster CMS on Linux Ubuntu 10.04 LTS v1.5 Global Open Versity e-Health Management Systems Hands-on Labs Training Manual Step-by-Step Install Guide OSCAR McMaster CMS on Linux Ubuntu 10.04 LTS (Lucid Lynx) Kefa Rabah Global Open Versity, Vancouver Canada [email protected] www.globalopenversity.org Table of Contents Page No. STEP-BY-STEP INSTALL GUIDE OSCAR MCMASTER CMS ON LINUX UBUNTU 10.04 LTS 3 1.0 Introduction 3 Part 1: Install & Configure Linux Ubuntu 10.04 LTS Desktop 4 Step 1: Getting Started & Hardware Pre-requisites 4 Step 2: Install Linux Ubuntu 10.04 LTS 4 Step 3: Update Ubuntu 10.04 Operating Systems 4 Part 2: Installing OSCAR McMaster CMS on Linux Ubuntu 8.04 LTS Server 5 Step 1: Preamble 5 Part 3: Installing the Infrastructure Packages 6 Step 1: Install OpenSSH Package 6 Step 2: Install Sun-Java6-JDK Package 7 Step 3: Install Ant Package 8 Step 4: Install MySQL Database Server Package 8 Step 5: Install Apache Tomcat 6 Package 9 Step 6: Install Pretty Good Privacy (PGP) Package 9 Step 7: Install CVS 10 Step 8: Install PostgreSQL database server 10 Step 9: Install UNZIP Package 10 Step 10: Install Network Time Protocol (NTP) Service 11 Step 11: Reboot the server 11 Step 12: Configuring the Base Packages 11 Step 13: Configuring Tomcat6 12 Step 14: Secure Apache Tomcat6 using SSL 13 Part 5: Install OSCAR CMS Server 15 Step 1: Download & Install OSCAR 15 Step 2: Create OSCAR Database on MySQL 16 Part 6: Enabling New eCharts 18 Step 1: Enable eChart 18 Part 7: Drugref2/3 -

Electronic Health Record Security Guidance for Low-Resource Settings
Assessment Tool for Electronic Health Record Security Guidance for Low-Resource Settings April 2020 MEASURE Evaluation This publication was produced with the support of the United States Agency for International Development (USAID) under University of North Carolina the terms of MEASURE Evaluation cooperative agreement 123 West Franklin Street, Suite 330 AID-OAA-L-14-00004. MEASURE Evaluation is implemented by the Carolina Population Center, University of North Carolina Chapel Hill, NC 27516 USA at Chapel Hill in partnership with ICF International; John Phone: +1 919-445-9350 Snow, Inc.; Management Sciences for Health; Palladium; [email protected] and Tulane University. Views expressed are not necessarily those of USAID or the United States government. MS-20-195 www.measureevaluation.org ISBN: 978-1-64232-260-6 ACKNOWLEDGMENTS We thank the United States Agency for International Development (USAID) for its support of this work. We’d like to recognize the project management team and the technical support we received from Jason B. Smith, Manish Kumar, and David La’Vell Johnson of MEASURE Evaluation at the University of North Carolina at Chapel Hill (UNC); Herman Tolentino, Steven Yoon, Tadesse Wuhib, Eric-Jan Manders, Daniel Rosen, Carrie Preston, Nathan Volk, and Adebowale Ojo of the United States Centers for Disease Control and Prevention (CDC); Mark DeZalia at the President’s Emergency Plan for AIDS Relief (PEPFAR); and Nega Gebreyesus, Kristen Wares, and Jacob Buehler at USAID. The MEASURE Evaluation project, funded by USAID and PEPFAR, would like to thank the Monitoring and Evaluation Technical Support Program at the Makerere University School of Public Health in Uganda for its cooperation and support in testing this tool and providing valuable feedback and insight.