Duplication of 10Q24 Locus: Broadening the Clinical and Radiological Spectrum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Variants in WNT2B and BTRC Predict Melanoma Survival
ACCEPTED MANUSCRIPT Genetic Variants in WNT2B and BTRC Predict Melanoma Survival Qiong Shi1, 2, 3, 9, Hongliang Liu2, 3, 9, Peng Han2, 3, 4, 9, Chunying Li1, Yanru Wang2, 3, Wenting Wu5, Dakai Zhu6, Christopher I. Amos6, Shenying Fang7, Jeffrey E. Lee7, Jiali Han5, 8* and Qingyi Wei2, 3* 1Department of Dermatology, Xijing Hospital, Fourth Military Medical University, Xi’an, Shaanxi 710032, China; 2Duke Cancer Institute, Duke University Medical Center, Durham, NC 27710, USA, 3Department of Medicine, Duke University School of Medicine, Durham, NC 27710, USA, 4Department of Otorhinolaryngology Head and Neck Surgery, First Affiliated Hospital, Xi'an Jiaotong University College of Medicine, Xi'an, Shaanxi 710061, China; 5Department of Epidemiology, Fairbanks School of Public Health, Indiana University Melvin and Bren Simon Cancer Center, Indiana University, Indianapolis,MANUSCRIPT IN 46202, USA 6Community and Family Medicine, Geisel School of Medicine, Dartmouth College, Hanover, NH 03755, USA; 7Department of Surgical Oncology, The University of Texas M. D. Anderson Cancer Center, Houston, Texas 77030, USA. 8Channing Division of Network Medicine, Department of Medicine, Brigham and Women’s Hospital, Boston, MA 02115, USA 9These authors contributed equally to this work. ACCEPTED *Correspondence: Qingyi Wei, M.D., Ph.D., Duke Cancer Institute, Duke University Medical Center and Department of Medicine, Duke School of Medicine, 905 S LaSalle Street, Durham, NC 27710, USA, Tel.: (919) 660-0562, E-mail: [email protected] and Jiali Han, M.D., Ph.D., 1 _________________________________________________________________________________ This is the author's manuscript of the article published in final edited form as: Shi, Q., Liu, H., Han, P., Li, C., Wang, Y., Wu, W., … Wei, Q. -

Mouse Germ Line Mutations Due to Retrotransposon Insertions Liane Gagnier1, Victoria P
Gagnier et al. Mobile DNA (2019) 10:15 https://doi.org/10.1186/s13100-019-0157-4 REVIEW Open Access Mouse germ line mutations due to retrotransposon insertions Liane Gagnier1, Victoria P. Belancio2 and Dixie L. Mager1* Abstract Transposable element (TE) insertions are responsible for a significant fraction of spontaneous germ line mutations reported in inbred mouse strains. This major contribution of TEs to the mutational landscape in mouse contrasts with the situation in human, where their relative contribution as germ line insertional mutagens is much lower. In this focussed review, we provide comprehensive lists of TE-induced mouse mutations, discuss the different TE types involved in these insertional mutations and elaborate on particularly interesting cases. We also discuss differences and similarities between the mutational role of TEs in mice and humans. Keywords: Endogenous retroviruses, Long terminal repeats, Long interspersed elements, Short interspersed elements, Germ line mutation, Inbred mice, Insertional mutagenesis, Transcriptional interference Background promoter and polyadenylation motifs and often a splice The mouse and human genomes harbor similar types of donor site [10, 11]. Sequences of full-length ERVs can TEs that have been discussed in many reviews, to which encode gag, pol and sometimes env, although groups of we refer the reader for more in depth and general infor- LTR retrotransposons with little or no retroviral hom- mation [1–9]. In general, both human and mouse con- ology also exist [6–9]. While not the subject of this re- tain ancient families of DNA transposons, none view, ERV LTRs can often act as cellular enhancers or currently active, which comprise 1–3% of these genomes promoters, creating chimeric transcripts with genes, and as well as many families or groups of retrotransposons, have been implicated in other regulatory functions [11– which have caused all the TE insertional mutations in 13]. -

Substrate Trapping Proteomics Reveals Targets of the Trcp2
Substrate Trapping Proteomics Reveals Targets of the TrCP2/FBXW11 Ubiquitin Ligase Tai Young Kim,a,b* Priscila F. Siesser,a,b Kent L. Rossman,b,c Dennis Goldfarb,a,d Kathryn Mackinnon,a,b Feng Yan,a,b XianHua Yi,e Michael J. MacCoss,e Randall T. Moon,f Channing J. Der,b,c Michael B. Majora,b,d Department of Cell Biology and Physiology,a Lineberger Comprehensive Cancer Center,b Department of Pharmacology,c and Department of Computer Science,d University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA; Department of Genome Sciencese and Department of Pharmacology and HHMI,f University of Washington, Seattle, Washington, USA Defining the full complement of substrates for each ubiquitin ligase remains an important challenge. Improvements in mass spectrometry instrumentation and computation and in protein biochemistry methods have resulted in several new methods for ubiquitin ligase substrate identification. Here we used the parallel adapter capture (PAC) proteomics approach to study TrCP2/FBXW11, a substrate adaptor for the SKP1–CUL1–F-box (SCF) E3 ubiquitin ligase complex. The processivity of the ubiquitylation reaction necessitates transient physical interactions between FBXW11 and its substrates, thus making biochemi- cal purification of FBXW11-bound substrates difficult. Using the PAC-based approach, we inhibited the proteasome to “trap” ubiquitylated substrates on the SCFFBXW11 E3 complex. Comparative mass spectrometry analysis of immunopurified FBXW11 protein complexes before and after proteasome inhibition revealed 21 known and 23 putatively novel substrates. In focused studies, we found that SCFFBXW11 bound, polyubiquitylated, and destabilized RAPGEF2, a guanine nucleotide exchange factor that activates the small GTPase RAP1. -

Syndromic Ear Anomalies and Renal Ultrasounds
Syndromic Ear Anomalies and Renal Ultrasounds Raymond Y. Wang, MD*; Dawn L. Earl, RN, CPNP‡; Robert O. Ruder, MD§; and John M. Graham, Jr, MD, ScD‡ ABSTRACT. Objective. Although many pediatricians cific MCA syndromes that have high incidences of renal pursue renal ultrasonography when patients are noted to anomalies. These include CHARGE association, Townes- have external ear malformations, there is much confusion Brocks syndrome, branchio-oto-renal syndrome, Nager over which specific ear malformations do and do not syndrome, Miller syndrome, and diabetic embryopathy. require imaging. The objective of this study was to de- Patients with auricular anomalies should be assessed lineate characteristics of a child with external ear malfor- carefully for accompanying dysmorphic features, includ- mations that suggest a greater risk of renal anomalies. We ing facial asymmetry; colobomas of the lid, iris, and highlight several multiple congenital anomaly (MCA) retina; choanal atresia; jaw hypoplasia; branchial cysts or syndromes that should be considered in a patient who sinuses; cardiac murmurs; distal limb anomalies; and has both ear and renal anomalies. imperforate or anteriorly placed anus. If any of these Methods. Charts of patients who had ear anomalies features are present, then a renal ultrasound is useful not and were seen for clinical genetics evaluations between only in discovering renal anomalies but also in the diag- 1981 and 2000 at Cedars-Sinai Medical Center in Los nosis and management of MCA syndromes themselves. Angeles and Dartmouth-Hitchcock Medical Center in A renal ultrasound should be performed in patients with New Hampshire were reviewed retrospectively. Only pa- isolated preauricular pits, cup ears, or any other ear tients who underwent renal ultrasound were included in anomaly accompanied by 1 or more of the following: the chart review. -
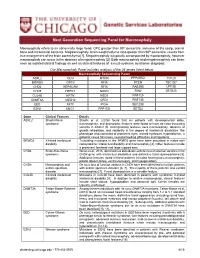
Macrocephaly Information Sheet 6-13-19
Next Generation Sequencing Panel for Macrocephaly Clinical Features: Macrocephaly refers to an abnormally large head, OFC greater than 98th percentile, inclusive of the scalp, cranial bone and intracranial contents. Megalencephaly, brain weight/volume ratio greater than 98th percentile, results from true enlargement of the brain parenchyma [1]. Megalencephaly is typically accompanied by macrocephaly, however macrocephaly can occur in the absence of megalencephaly [2]. Both macrocephaly and megalencephaly can been seen as isolated clinical findings as well as clinical features of a mutli-systemic syndromic diagnosis. Our Macrocephaly Panel includes analysis of the 36 genes listed below. Macrocephaly Sequencing Panel ASXL2 GLI3 MTOR PPP2R5D TCF20 BRWD3 GPC3 NFIA PTEN TBC1D7 CHD4 HEPACAM NFIX RAB39B UPF3B CHD8 HERC1 NONO RIN2 ZBTB20 CUL4B KPTN NSD1 RNF125 DNMT3A MED12 OFD1 RNF135 EED MITF PIGA SEC23B EZH2 MLC1 PPP1CB SETD2 Gene Clinical Features Details ASXL2 Shashi-Pena Shashi et al. (2016) found that six patients with developmental delay, syndrome macrocephaly, and dysmorphic features were found to have de novo truncating variants in ASXL2 [3]. Distinguishing features were macrocephaly, absence of growth retardation, and variability in the degree of intellectual disabilities The phenotype also consisted of prominent eyes, arched eyebrows, hypertelorism, a glabellar nevus flammeus, neonatal feeding difficulties and hypotonia. BRWD3 X-linked intellectual Truncating mutations in the BRWD3 gene have been described in males with disability nonsyndromic intellectual disability and macrocephaly [4]. Other features include a prominent forehead and large cupped ears. CHD4 Sifrim-Hitz-Weiss Weiss et al., 2016, identified five individuals with de novo missense variants in the syndrome CHD4 gene with intellectual disabilities and distinctive facial dysmorphisms [5]. -

Supplementary Table 1. Genes Mapped in Core Cancer
Supplementary Table 1. Genes mapped in core cancer pathways annotated by KEGG (Kyoto Encyclopedia of Genes and Genomes), MIPS (The Munich Information Center for Protein Sequences), BIOCARTA, PID (Pathway Interaction Database), and REACTOME databases. EP300,MAP2K1,APC,MAP3K7,ZFYVE9,TGFB2,TGFB1,CREBBP,MAP BIOCARTA TGFB PATHWAY K3,TAB1,SMAD3,SMAD4,TGFBR2,SKIL,TGFBR1,SMAD7,TGFB3,CD H1,SMAD2 TFDP1,NOG,TNF,GDF7,INHBB,INHBC,COMP,INHBA,THBS4,RHOA,C REBBP,ROCK1,ID1,ID2,RPS6KB1,RPS6KB2,CUL1,LOC728622,ID4,SM AD3,MAPK3,RBL2,SMAD4,RBL1,NODAL,SMAD1,MYC,SMAD2,MAP K1,SMURF2,SMURF1,EP300,BMP8A,GDF5,SKP1,CHRD,TGFB2,TGFB 1,IFNG,CDKN2B,PPP2CB,PPP2CA,PPP2R1A,ID3,SMAD5,RBX1,FST,PI KEGG TGF BETA SIGNALING PATHWAY TX2,PPP2R1B,TGFBR2,AMHR2,LTBP1,LEFTY1,AMH,TGFBR1,SMAD 9,LEFTY2,SMAD7,ROCK2,TGFB3,SMAD6,BMPR2,GDF6,BMPR1A,B MPR1B,ACVRL1,ACVR2B,ACVR2A,ACVR1,BMP4,E2F5,BMP2,ACVR 1C,E2F4,SP1,BMP7,BMP8B,ZFYVE9,BMP5,BMP6,ZFYVE16,THBS3,IN HBE,THBS2,DCN,THBS1, JUN,LRP5,LRP6,PPP3R2,SFRP2,SFRP1,PPP3CC,VANGL1,PPP3R1,FZD 1,FZD4,APC2,FZD6,FZD7,SENP2,FZD8,LEF1,CREBBP,FZD9,PRICKLE 1,CTBP2,ROCK1,CTBP1,WNT9B,WNT9A,CTNNBIP1,DAAM2,TBL1X R1,MMP7,CER1,MAP3K7,VANGL2,WNT2B,WNT11,WNT10B,DKK2,L OC728622,CHP2,AXIN1,AXIN2,DKK4,NFAT5,MYC,SOX17,CSNK2A1, CSNK2A2,NFATC4,CSNK1A1,NFATC3,CSNK1E,BTRC,PRKX,SKP1,FB XW11,RBX1,CSNK2B,SIAH1,TBL1Y,WNT5B,CCND1,CAMK2A,NLK, CAMK2B,CAMK2D,CAMK2G,PRKACA,APC,PRKACB,PRKACG,WNT 16,DAAM1,CHD8,FRAT1,CACYBP,CCND2,NFATC2,NFATC1,CCND3,P KEGG WNT SIGNALING PATHWAY LCB2,PLCB1,CSNK1A1L,PRKCB,PLCB3,PRKCA,PLCB4,WIF1,PRICK LE2,PORCN,RHOA,FRAT2,PRKCG,MAPK9,MAPK10,WNT3A,DVL3,R -

Deregulated Wnt/Β-Catenin Program in High-Risk Neuroblastomas Without
Oncogene (2008) 27, 1478–1488 & 2008 Nature Publishing Group All rights reserved 0950-9232/08 $30.00 www.nature.com/onc ONCOGENOMICS Deregulated Wnt/b-catenin program in high-risk neuroblastomas without MYCN amplification X Liu1, P Mazanek1, V Dam1, Q Wang1, H Zhao2, R Guo2, J Jagannathan1, A Cnaan2, JM Maris1,3 and MD Hogarty1,3 1Division of Oncology, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA; 2Department of Biostatistics and Epidemiology, University of Pennsylvania School of Medicine, Philadelphia, PA, USA and 3Department of Pediatrics, University of Pennsylvania School of Medicine, Philadelphia, PA, USA Neuroblastoma (NB) is a frequently lethal tumor of Introduction childhood. MYCN amplification accounts for the aggres- sive phenotype in a subset while the majority have no Neuroblastoma (NB) is a childhood embryonal malig- consistently identified molecular aberration but frequently nancy arising in the peripheral sympathetic nervous express MYC at high levels. We hypothesized that acti- system. Half of all children with NB present with features vated Wnt/b-catenin (CTNNB1) signaling might account that define their tumorsashigh riskwith poor overall for this as MYC is a b-catenin transcriptional target and survival despite intensive therapy (Matthay et al., 1999). multiple embryonal and neural crest malignancies have A subset of these tumors are characterized by high-level oncogenic alterations in this pathway. NB cell lines without genomic amplification of the MYCN proto-oncogene MYCN amplification express higher levels of MYC and (Matthay et al., 1999) but the remainder have no b-catenin (with aberrant nuclear localization) than MYCN- consistently identified aberration to account for their amplified cell lines. -

Polydactyly of the Hand
A Review Paper Polydactyly of the Hand Katherine C. Faust, MD, Tara Kimbrough, BS, Jean Evans Oakes, MD, J. Ollie Edmunds, MD, and Donald C. Faust, MD cleft lip/palate, and spina bifida. Thumb duplication occurs in Abstract 0.08 to 1.4 per 1000 live births and is more common in Ameri- Polydactyly is considered either the most or second can Indians and Asians than in other races.5,10 It occurs in a most (after syndactyly) common congenital hand ab- male-to-female ratio of 2.5 to 1 and is most often unilateral.5 normality. Polydactyly is not simply a duplication; the Postaxial polydactyly is predominant in black infants; it is most anatomy is abnormal with hypoplastic structures, ab- often inherited in an autosomal dominant fashion, if isolated, 1 normally contoured joints, and anomalous tendon and or in an autosomal recessive pattern, if syndromic. A prospec- ligament insertions. There are many ways to classify tive San Diego study of 11,161 newborns found postaxial type polydactyly, and surgical options range from simple B polydactyly in 1 per 531 live births (1 per 143 black infants, excision to complicated bone, ligament, and tendon 1 per 1339 white infants); 76% of cases were bilateral, and 3 realignments. The prevalence of polydactyly makes it 86% had a positive family history. In patients of non-African descent, it is associated with anomalies in other organs. Central important for orthopedic surgeons to understand the duplication is rare and often autosomal dominant.5,10 basic tenets of the abnormality. Genetics and Development As early as 1896, the heritability of polydactyly was noted.11 As olydactyly is the presence of extra digits. -

Familial Poland Anomaly
J Med Genet: first published as 10.1136/jmg.19.4.293 on 1 August 1982. Downloaded from Journal ofMedical Genetics, 1982, 19, 293-296 Familial Poland anomaly T J DAVID From the Department of Child Health, University of Manchester, Booth Hall Children's Hospital, Manchester SUMMARY The Poland anomaly is usually a non-genetic malformation syndrome. This paper reports two second cousins who both had a typical left sided Poland anomaly, and this constitutes the first recorded case of this condition affecting more than one member of a family. Despite this, for the purposes of genetic counselling, the Poland anomaly can be regarded as a sporadic condition with an extremely low recurrence risk. The Poland anomaly comprises congenital unilateral slightly reduced. The hands were normal. Another absence of part of the pectoralis major muscle in son (Greif himself) said that his own left pectoralis combination with a widely varying spectrum of major was weaker than the right. "Although the ipsilateral upper limb defects.'-4 There are, in difference is obvious, the author still had to carry addition, patients with absence of the pectoralis out his military duties"! major in whom the upper limbs are normal, and Trosev and colleagues9 have been widely quoted as much confusion has been caused by the careless reporting familial cases of the Poland anomaly. labelling of this isolated defect as the Poland However, this is untrue. They described a mother anomaly. It is possible that the two disorders are and child with autosomal dominant radial sided part of a single spectrum, though this has never been upper limb defects. -

A Model for Human Ectrodactyly SHFM3
Article Characterization of mouse Dactylaplasia mutations: a model for human ectrodactyly SHFM3 FRIEDLI, Marc, et al. Abstract SHFM3 is a limb malformation characterized by the absence of central digits. It has been shown that this condition is associated with tandem duplications of about 500 kb at 10q24. The Dactylaplasia mice display equivalent limb defects and the two corresponding alleles (Dac1j and Dac2j) map in the region syntenic with the duplications in SHFM3. Dac1j was shown to be associated with an insertion of an unspecified ETn-like mouse endogenous transposon upstream of the Fbxw4 gene. Dac2j was also thought to be an insertion or a small inversion in intron 5 of Fbxw4, but the breakpoints and the exact molecular lesion have not yet been characterized. Here we report precise mapping and characterization of these alleles. We failed to identify any copy number differences within the SHFM3 orthologous genomic locus between Dac mutant and wild-type littermates, showing that the Dactylaplasia alleles are not associated with duplications of the region, in contrast with the described human SHFM3 cases. We further show that both Dac1j and Dac2j are caused by insertions of MusD retroelements that share 98% sequence identity. The differences [...] Reference FRIEDLI, Marc, et al. Characterization of mouse Dactylaplasia mutations: a model for human ectrodactyly SHFM3. Mammalian Genome, 2008, vol. 19, no. 4, p. 272-8 PMID : 18392654 DOI : 10.1007/s00335-008-9106-0 Available at: http://archive-ouverte.unige.ch/unige:1039 Disclaimer: layout of this document may differ from the published version. 1 / 1 Mamm Genome (2008) 19:272–278 DOI 10.1007/s00335-008-9106-0 Characterization of mouse Dactylaplasia mutations: a model for human ectrodactyly SHFM3 Marc Friedli Æ Sergey Nikolaev Æ Robert Lyle Æ Me´lanie Arcangeli Æ Denis Duboule Æ Franc¸ois Spitz Æ Stylianos E. -

Regulation of BTRC by Gain-Of-Function TP53 Mutation(S) in Cancer Cells
Regulation of BTRC by gain-of-function TP53 mutation(s) in cancer cells By Peter Xu A thesis submitted in conformity with the requirements for the degree of Masters of Science (M.Sc) Graduate Department of Molecular Genetics University of Toronto © Copyright by Peter Xu (2015) Regulation of BTRC by gain-of-function TP53 mutation(s) in cancer cells Peter Xu Masters of Science Department of Molecular Genetics University of Toronto 2015 ABSTRACT Regulation of BTRC by gain-of-function TP53 mutation Peter Xu, Masters of Science (2015), Department of Molecular Genetics, University of Toronto Mutation or loss of TP53 has been detected in more than 50% of all human tumours. Gain-of-function (GOF) TP53 mutations have been linked to metastasis, altered metabolism, and drug resistance. Cancer patients carrying GOF TP53 mutations in their tumours respond poorly to standard of care treatments and have a worse prognosis. In my Master’s thesis, I have focused on understanding the relationship between a frequently observed mutation of TP53 (TP53 R248W ), and BTRC , an E3 ubiquitin ligase with functions impinging on cell cycle, morphology and metabolism. I observed a correlation between BTRC protein expression and TP53 genotype across a panel cancer cell lines. Additionally, I observed that TP53 R248W -mediated regulation of BTRC expression is dependent on a post-transcriptional mechanism, likely involving more than one micro-RNA. In conclusion, I present a model describing the regulation of BTRC by TP53 , which may have implications for targeted strategies in cancers harboring GOF TP53 mutations. ii ACKNOWLEDGEMENTS Foremost, I would like to express my sincere gratitude to my advisor Prof. -

An Integrated Genome Screen Identifies the Wnt Signaling Pathway As a Major Target of WT1
An integrated genome screen identifies the Wnt signaling pathway as a major target of WT1 Marianne K.-H. Kima,b, Thomas J. McGarryc, Pilib O´ Broind, Jared M. Flatowb, Aaron A.-J. Goldend, and Jonathan D. Lichta,b,1 aDivision of Hematology/Oncology and cFeinberg Cardiovascular Research Institute, Division of Cardiology, Northwestern University Feinberg School of Medicine, Chicago, IL 60611; bRobert H. Lurie Cancer Center, Northwestern University, Chicago, IL 60611; and dDepartment of Information Technology, National University of Ireland, Galway, Republic of Ireland Edited by Peter K. Vogt, The Scripps Research Institute, La Jolla, CA, and approved May 18, 2009 (received for review February 12, 2009) WT1, a critical regulator of kidney development, is a tumor suppressor control. A first generation of in vitro cotransfection and DNA for nephroblastoma but in some contexts functions as an oncogene. binding assays have given way to more biologically relevant exper- A limited number of direct transcriptional targets of WT1 have been iments where manipulation of WT1 levels has been accompanied identified to explain its complex roles in tumorigenesis and organo- by examination of candidate or global gene expression. Classes of genesis. In this study we performed genome-wide screening for direct WT1 targets thus identified consist of inducers of differentiation, WT1 targets, using a combination of ChIP–ChIP and expression arrays. regulators of cell growth and modulators of cell death. WT1 target Promoter regions bound by WT1 were highly G-rich and resembled genes include CDKN1A (22), a negative regulator of the cell cycle; the sites for a number of other widely expressed transcription factors AREG (23), a facilitator of kidney differentiation; WNT4 (24), a such as SP1, MAZ, and ZNF219.