Cytokine Production by Suppressing Tollip Promotes TLR-Triggered
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
![RT² Profiler PCR Array (96-Well Format and 384-Well [4 X 96] Format)](https://docslib.b-cdn.net/cover/6983/rt%C2%B2-profiler-pcr-array-96-well-format-and-384-well-4-x-96-format-616983.webp)
RT² Profiler PCR Array (96-Well Format and 384-Well [4 X 96] Format)
RT² Profiler PCR Array (96-Well Format and 384-Well [4 x 96] Format) Human Toll-Like Receptor Signaling Pathway Cat. no. 330231 PAHS-018ZA For pathway expression analysis Format For use with the following real-time cyclers RT² Profiler PCR Array, Applied Biosystems® models 5700, 7000, 7300, 7500, Format A 7700, 7900HT, ViiA™ 7 (96-well block); Bio-Rad® models iCycler®, iQ™5, MyiQ™, MyiQ2; Bio-Rad/MJ Research Chromo4™; Eppendorf® Mastercycler® ep realplex models 2, 2s, 4, 4s; Stratagene® models Mx3005P®, Mx3000P®; Takara TP-800 RT² Profiler PCR Array, Applied Biosystems models 7500 (Fast block), 7900HT (Fast Format C block), StepOnePlus™, ViiA 7 (Fast block) RT² Profiler PCR Array, Bio-Rad CFX96™; Bio-Rad/MJ Research models DNA Format D Engine Opticon®, DNA Engine Opticon 2; Stratagene Mx4000® RT² Profiler PCR Array, Applied Biosystems models 7900HT (384-well block), ViiA 7 Format E (384-well block); Bio-Rad CFX384™ RT² Profiler PCR Array, Roche® LightCycler® 480 (96-well block) Format F RT² Profiler PCR Array, Roche LightCycler 480 (384-well block) Format G RT² Profiler PCR Array, Fluidigm® BioMark™ Format H Sample & Assay Technologies Description The Human Toll-Like Receptor (TLR) Signaling Pathway RT² Profiler PCR Array profiles the expression of 84 genes central to TLR-mediated signal transduction and innate immunity. The TLR family of pattern recognition receptors (PRRs) detects a wide range of bacteria, viruses, fungi and parasites via pathogen-associated molecular patterns (PAMPs). Each receptor binds to specific ligands, initiates a tailored innate immune response to the specific class of pathogen, and activates the adaptive immune response. -

TLR Signaling Pathways
Seminars in Immunology 16 (2004) 3–9 TLR signaling pathways Kiyoshi Takeda, Shizuo Akira∗ Department of Host Defense, Research Institute for Microbial Diseases, Osaka University, and ERATO, Japan Science and Technology Corporation, 3-1 Yamada-oka, Suita, Osaka 565-0871, Japan Abstract Toll-like receptors (TLRs) have been established to play an essential role in the activation of innate immunity by recognizing spe- cific patterns of microbial components. TLR signaling pathways arise from intracytoplasmic TIR domains, which are conserved among all TLRs. Recent accumulating evidence has demonstrated that TIR domain-containing adaptors, such as MyD88, TIRAP, and TRIF, modulate TLR signaling pathways. MyD88 is essential for the induction of inflammatory cytokines triggered by all TLRs. TIRAP is specifically involved in the MyD88-dependent pathway via TLR2 and TLR4, whereas TRIF is implicated in the TLR3- and TLR4-mediated MyD88-independent pathway. Thus, TIR domain-containing adaptors provide specificity of TLR signaling. © 2003 Elsevier Ltd. All rights reserved. Keywords: TLR; Innate immunity; Signal transduction; TIR domain 1. Introduction 2. Toll-like receptors Toll receptor was originally identified in Drosophila as an A mammalian homologue of Drosophila Toll receptor essential receptor for the establishment of the dorso-ventral (now termed TLR4) was shown to induce the expression pattern in developing embryos [1]. In 1996, Hoffmann and of genes involved in inflammatory responses [3]. In addi- colleagues demonstrated that Toll-mutant flies were highly tion, a mutation in the Tlr4 gene was identified in mouse susceptible to fungal infection [2]. This study made us strains that were hyporesponsive to lipopolysaccharide [4]. aware that the immune system, particularly the innate im- Since then, Toll receptors in mammals have been a major mune system, has a skilful means of detecting invasion by focus in the immunology field. -

Innate Immunity and Inflammation of the Bovine Female Reproductive Tract
REPRODUCTIONREVIEW Innate immunity and inflammation of the bovine female reproductive tract in health and disease I Martin Sheldon, James G Cronin, Gareth D Healey, Christoph Gabler1, Wolfgang Heuwieser2, Dominik Streyl3, John J Bromfield4, Akio Miyamoto5, Chrys Fergani6 and Hilary Dobson6 College of Medicine, Institute of Life Science, Swansea University, Singleton Park, Swansea SA2 8PP, UK, 1Institute of Veterinary Biochemistry and 2Clinic of Animal Reproduction, Freie Universitaet Berlin, Berlin, Germany, 3Clinic for Ruminants with Ambulatory and Herd Health Services, Centre for Clinical Veterinary Medicine, Ludwig Maximilian University of Munich, Oberschleißheim, Germany, 4Department of Animal Sciences, University of Florida, Gainesville, Florida 32608, USA, 5Graduate School for Animal and Food Hygiene, Obihiro University of Agriculture and Veterinary Medicine, Obihiro 080-8555, Japan and 6School of Veterinary Science, University of Liverpool, Leahurst, Neston CH64 7TE, UK Correspondence should be addressed to I M Sheldon; Email: [email protected] Abstract Mammalian reproductive physiology and the development of viviparity co-evolved with inflammation and immunity over millennia. Many inflammatory mediators contribute to paracrine and endocrine signalling, and the maintenance of tissue homeostasis in the female reproductive tract. However, inflammation is also a feature of microbial infections of the reproductive tract. Bacteria and viruses commonly cause endometritis, perturb ovarian follicle development and suppress the endocrine activity of the hypothalamus and pituitary in cattle. Innate immunity is an evolutionary ancient system that orchestrates host cell inflammatory responses aimed at eliminating pathogens and repairing damaged tissue. Pattern recognition receptors on host cells bind pathogen-associated molecular patterns and damage-associated molecular patterns, leading to the activation of intracellular MAPK and NFkB signalling pathways and the release of inflammatory mediators. -

Inflammation in Innate and Adaptive Immune Mechanisms October 28 – 30, 2012 Hilton Grand Wailea Resort | Maui, Hawai’I Organizers: Tom Hamilton and Xiaoxia Li
45th Annual Meeting of The Society for Leukocyte Biology InflammatIon In Innate and adaptIve Immune mechanIsms October 28 – 30, 2012 Hilton Grand Wailea Resort | Maui, Hawai’i Organizers: Tom Hamilton and Xiaoxia Li abstracts Journal of Leukocyte Biology, Supplement 2012 www.leukocytebiology.org 45th AnnuAl Meeting of the Society for leukocyte Biology inflAMMAtion in innAte And AdAptive iMMune MechAniSMS grAnd WAileA, MAui, hAWAi’i ~ octoBer 28-30, 2012 Thank you to the supporters of the 2012 SLB Meeting! Journal of Leukocyte Biology Supplement 2012 ABSTRACTS 1 2 Alcohol and Drugs of Abuse Interaction with HIV/AIDS: Chronic Alcohol Consumption Increases Mortality in Sepsis Systems Biology Approach in the SIV-Infected Macaque Benyam P. Yoseph, Zhe Liang, Elise Breed, Kevin McConnell, Patricia E. Molina David M. Guidot, Michael Koval, Craig M. Coopersmith Comprehensive Alcohol Research Center, Louisiana State Emory University School of Medicine University Health Sciences Center, NOLA Introduction: Excessive alcohol abuse is a problem of particular The two most commonly used and abused drugs are alcohol concern in the intensive care unit (ICU), as the rate of morbidity and the cannabinoids. Alcohol and drugs of abuse have been and mortality in all patients admitted to ICU is 2-4 times greater demonstrated to alter host response to human immunodeficiency than in non-alcoholics. Sepsis is the leading cause of death in ICU. (HIV) infection; by affecting progression of infection, tissue The purpose of this study was to examine the pathophysiology of injury, and time to death. Several factors can be involved in chronic alcohol abuse in sepsis. this, those pertaining to the host response, as well as those Methods: FVB/N mice were given liquid ethanol diet (20% w/v) or related to the ability of the virus to integrate itself into the host water for 12 weeks. -

Immune Stress up Regulates TLR4 and Tollip Gene Expression in the Hypothalamus of Ewes
Journal of Animal and Feed Sciences, Vol. 22, No. 1, 2013, 13–18 The Kielanowski Institute of Animal Physiology and Nutrition PAS Jabłonna Immune stress up regulates TLR4 and Tollip gene expression in the hypothalamus of ewes A.P. Herman1,3, A. Herman2, K. Haziak1, and D. Tomaszewska-Zaremba1 1 The Kielanowski Institute of Animal Physiology and Nutrition, Polish Academy of Sciences, 05-110 Jabłonna, Poland 2 The Academy of Cosmetics and Health Care in Warsaw, 00-252 Warsaw, Poland KEY WORDS: ewe, hypothalamus, ABSTRACT. Bacterial endotoxin, LPS, is recognized by Toll-like receptor-4 lipopolysaccharide, Toll-like receptor-4, (TLR4) and induces a signaling cascade leading to synthesis of proinflamma- Toll-interacting protein tory cytokines and induction of sickness behavior in animals. Transduction of the TLR4 signal is controlled by a potent negative regulator—Toll-interacting protein. The presented study concerns the effect of intravenously injected LPS on the level of expression of TLR4 and Tollip genes in the hypothalamus of ewes. Endotoxin increased (P < 0.01) cortisol release and expression of TLR4 and Tollip genes in the preoptic area (1.87 ± 0.42 and 1.31 ± 0.15), Received 19 September 2012 anterior hypothalamus (1.77 ± 0.22 and 1.27 ± 0.13), medial basal hypothala- Revised 18 February 2013 mus (2.53 0.65 and 1.43 ± 0.15), and median eminence (2.93 ± 0.46 and Accepted 18 March 2013 1.73 ± 0.10), respectively, in comparison with non-treated animals. Our results show that immune stress increases TLR4 gene expression in the hypothala- 3 Corresponding author: mus. -

Inflammation Fas Modulating Intestinal TLR-Mediated Through TLR4 And
Intestinal Expression of Fas and Fas Ligand Is Upregulated by Bacterial Signaling through TLR4 and TLR5, with Activation of Fas Modulating Intestinal TLR-Mediated This information is current as Inflammation of September 24, 2021. Philana Fernandes, Charlotte O'Donnell, Caitriona Lyons, Jonathan Keane, Tim Regan, Stephen O'Brien, Padraic Fallon, Elizabeth Brint and Aileen Houston J Immunol 2014; 193:6103-6113; Prepublished online 5 Downloaded from November 2014; doi: 10.4049/jimmunol.1303083 http://www.jimmunol.org/content/193/12/6103 http://www.jimmunol.org/ Supplementary http://www.jimmunol.org/content/suppl/2014/11/05/jimmunol.130308 Material 3.DCSupplemental References This article cites 43 articles, 20 of which you can access for free at: http://www.jimmunol.org/content/193/12/6103.full#ref-list-1 by guest on September 24, 2021 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD -

TOLLIP Promotes Durable Alveolar Macrophage-Mediated Immunity During Mycobacterium Tuberculosis Infection by Resolving Cellular
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.24.263624; this version posted August 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 TOLLIP promotes durable alveolar macrophage-mediated immunity during Mycobacterium 2 tuberculosis infection by resolving cellular stress from lipids. 3 4 Sambasivan Venkatasubramanian1, Courtney Plumlee2, Kim Dill-McFarland1, Gemma L. 5 Pearson3, Sara B. Cohen2, Anne Lietzke3, Amanda Pacheco3, Robyn Pryor1, Scott A. 6 Soleimanpour3,4, Matthew Altman1, Kevin B. Urdahl2,5, Javeed A. Shah1,6*. 7 1 Department of Medicine, University of Washington, Seattle, WA. 8 2 Seattle Children’s Research Institute, Seattle, WA. 9 3 Department of Internal Medicine, University of Michigan, Ann Arbor, MI. 10 4 VA Ann Arbor Healthcare System, Ann Arbor, MI. 11 5 Departments of Pediatrics and Immunology, University of Washington, Seattle, WA. 12 6 VA Puget Sound Healthcare System, Seattle, WA. * Correspondence: [email protected]; @ShahLab. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.08.24.263624; this version posted August 24, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 13 Summary 14 TOLLIP, a ubiquitin binding protein that controls multiple macrophage functions via 15 endoplasmic reticulum transport and autophagy, is associated with human tuberculosis (TB) 16 susceptibility and immune responses in genetic studies. -

Product Data Sheet
KAMIYA BIOMEDICAL COMPANY Rev. 091305 PRODUCT DATA SHEET Product: Toll Interacting Protein (TOLLIP/TOLIP) (IN) polyclonal Cat. No.: PC-599 (100 µµµg) Storage and Stability : Store at -20˚C. Aliquot to avoid repeat Background: freeze/thaw cycles. Toll-like receptors (TRLs) are evolutionarily conserved pattern recognition molecules Applications and Suggested Dilutions: resembling the toll proteins that mediate Western blot: use at 1 µg/mL antimicrobial responses in Drosophila. These Immunohistochemistry proteins recognize different microbial products during infection and serve as an important link The optimal dilution for a specific application between the innate and adaptive immune should be determined by the researcher. responses. The TLRs act through adaprot molecules to activate various kinases and Limitations: transcription factors so the organism can For in vitro research use only. Not for use in respond to potential infection. These adaptor diagnostics or in humans. molecules include TOLLIP, MyD88, and TRIF. TOLLIP associates directly with TLR2 and TLR4, Warranty: acting as an inhibitor to TLR activation. This No warranties, expressed or implied, are made negative regulation of TLR signaling may serve regarding the use of this product. KAMIYA to limit the production of pro-inflammatory BIOMEDICAL COMPANY is not liable for any mediators during infection and inflammation. damage, personal injury, or economic loss caused by this product. Molecular Weight: 30 kD kDa Species Reactivity: Human, Rat, and Mouse Host : Rabbit Isotype: IgG Positive Control: Rat brain lysate. Located in cytoplasm and phosphorylated by IRAK1 upon stimulation by IL-1 or microbial products. Immunogen: Rabbit polyclonal TOLLIP antibody was raised against a peptide corresponding to 16 amino acids near the center of human TOLLIP. -

Lipopolysaccharide Signaling in Endothelial Cells
Laboratory Investigation (2006) 86, 9–22 & 2006 USCAP, Inc All rights reserved 0023-6837/06 $30.00 www.laboratoryinvestigation.org Lipopolysaccharide signaling in endothelial cells Shauna M Dauphinee1,2 and Aly Karsan1,2,3,4 1Department of Medical Biophysics, British Columbia Cancer Agency, Vancouver, British Columbia, Canada; 2Experimental Medicine Program, University of British Columbia, Vancouver, British Columbia, Canada; 3Department of Pathology and Laboratory Medicine, British Columbia Cancer Agency, Vancouver, British Columbia, Canada and 4Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada Sepsis is the systemic immune response to severe bacterial infection. The innate immune recognition of bacterial and viral products is mediated by a family of transmembrane receptors known as Toll-like receptors (TLRs). In endothelial cells, exposure to lipopolysaccharide (LPS), a major cell wall constituent of Gram- negative bacteria, results in endothelial activation through a receptor complex consisting of TLR4, CD14 and MD2. Recruitment of the adaptor protein myeloid differentiation factor (MyD88) initiates an MyD88-dependent pathway that culminates in the early activation of nuclear factor-jB (NF-jB) and the mitogen-activated protein kinases. In parallel, a MyD88-independent pathway results in a late-phase activation of NF-jB. The outcome is the production of various proinflammatory mediators and ultimately cellular injury, leading to the various vascular sequelae of sepsis. -

Functional Expression of TLR5 of Different Vertebrate Species And
www.nature.com/scientificreports OPEN Functional expression of TLR5 of diferent vertebrate species and diversifcation in intestinal Received: 4 December 2017 Accepted: 10 July 2018 pathogen recognition Published: xx xx xxxx Eugenia Faber1,2, Karsten Tedin3, Yvonne Speidel1,2, Melanie M. Brinkmann4 & Christine Josenhans 1,2,5 Toll-like receptor 5 (TLR5) is activated by bacterial fagellins and plays a crucial role in the frst-line defence against pathogenic bacteria and in immune homeostasis, and is highly conserved in vertebrate species. However, little comparative information is available on TLR5 functionality. In this study, we compared TLR5 activation using full-length and chimeric TLR5 of various vertebrate species (human, chicken, mouse, pig, cattle). Chimeric TLR5 receptors, consisting of human transmembrane and intracellular domains, linked to extracellular domains of animal origin, were generated and expressed. The comparison of chimeric TLR5s and their full-length counterparts revealed signifcant functional disparities. While porcine and chicken full-length TLR5s showed a strongly reduced functionality in human cells, all chimeric receptors were functional when challenged with TLR5 ligand Salmonella FliC. Using chimeric receptors as a tool allowed for the identifcation of ectodomain-dependent activation potential and partially host species-specifc diferences in response to various enteric bacterial strains and their purifed fagellins. We conclude that both the extra- and intracellular determinants of TLR5 receptors are crucial for compatibility with the species expression background and hence for proper receptor functionality. TLR5 receptors with a common intracellular domain provide a useful system to investigate bacteria- and host-specifc diferences in receptor activation. Toll-like receptor 5 (TLR5) is a crucial determinant of pathogen-host interaction and essential for immune home- ostasis1–4. -

Tollip-Induced Down-Regulation of MARCH1
Results in Immunology 3 (2013) 17–25 Contents lists available at ScienceDirect Results in Immunology j o u r n a l h o m e p a g e : w w w . e l s e v i e r . c o m / l o c a t e / r i n i m ଝ Tollip-induced down-regulation of MARCH1 Marie-Claude Bourgeois-Daigneault a, Abdul Mohammad Pezeshki a, Tristan Galbas a, Mathieu Houde b, Martin Baril b, Klaus Fr uh¨ c, Abdelaziz Amrani d, Satoshi Ishido e, Daniel Lamarre b, Jacques Thibodeau a,* aLaboratoire d’Immunologie Mol ´eculaire, D ´epartement de Microbiologie et Immunologie, Universit ´e de Montr ´eal, QC, Canada bInstitut de recherche en immunologie et en canc ´erologie, IRIC, Universit ´e de Montr ´eal, QC, Canada cVaccine and Gene Therapy Institute, Oregon Health and Science University, Beaverton, OR, USA dD ´epartement de P ´ediatrie, Service d ’Immunologie et d’Allergologie, Universit ´e de Sherbrooke, Sherbrooke, QC, Canada eLaboratory of Infectious Immunity, Yokohama Institute, Research Center for Allergy and Immunology, Japan a r t i c l e i n f o a b s t r a c t Article history: In addition to their classical antigen presenting functions, MHC class II molecules potentiate the TLR-triggered Received 29 January 2013 production of pro-inflammatory cytokines. Here, we have addressed the effect of Tollip and MARCH1 on the Received in revised form 13 February 2013 regulation of MHC II trafficking and TLR signaling. Our results show that MARCH1-deficient mice splenocytes Accepted 14 February 2013 are impaired in their capacity to produce pro-inflammatory cytokines in response to poly(I:C) and that TLR3 and MHC II molecules interact in the endocytic pathway. -
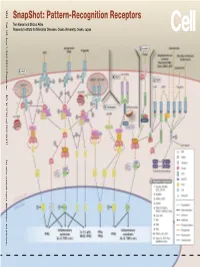
Snapshot: Pattern-Recognition Receptors
SnapShot: Pattern-Recognition Receptors SnapShot:Pattern-Recognition Kawai and Shizuo Akira Taro Osaka, Japan Diseases, Osaka University, Institute for Microbial Research 1024 Cell 129, June 1, 2007 ©2007 Elsevier Inc. DOI 10.1016/j.cell.2007.05.017 See online version for legend, abbreviations, and references. SnapShot: Pattern-Recognition Receptors Taro Kawai and Shizuo Akira Research Institute for Microbial Diseases, Osaka University, Osaka, Japan (A) Toll-like receptor signaling. Toll-like receptor (TLR) 3 recognizes polyinosinic-polycytidylic acid (poly IC), whereas TLR4 recognizes lipopolysaccharide (LPS). TLR2 recog- nizes various components such as lipoprotein and peptidoglycan (PGN). TLR5 detects flagellin. TLR7 and TLR9 detect single-stranded (ss)RNA and CpG DNA, respectively. Each TLR recruits a distinct set of Toll/interleukin-1 receptor (TIR) domain-containing adaptor molecules such as myeloid differentiation primary response gene 88 (MyD88), TIR-containing adaptor protein (TIRAP, also known as MAL), TIR-containing adaptor-inducing IFNβ (TRIF, also known as TICAM1) and TRIF-related adaptor molecule (TRAM, also known as TICAM2). TLR3 uses TRIF, and TLR5, 7, and 9 use MyD88. TLR2 uses MyD88 and TIRAP, and TLR4 uses MyD88, TIRAP, TRIF, and TRAM. MyD88 binds to inter- leukin-1 receptor-associated kinase 4 (IRAK4) and TRAF6. TRIF binds receptor-interacting protein 1 (RIP1) and TRAF6. TRAF6 forms a complex with Ubc13, Uev1A, and ECSIT (evolutionarily conserved signaling intermediate in Toll/IL-1R pathways) to activate a complex containing transforming growth factor-β-activated kinase 1 (TAK1), TAK1-binding protein 1 (TAB1), TAB2, and TAB3. TAK1 activates IκB kinase (IKK) complex consisting of IKKα, IKKβ, and Nemo (also known as IKKγ), which results in the phosphorylation and proteasomal degradation of IκB proteins and the release of a transcription factor NFκB to the nucleus to regulate expression of inflammatory cytokines such as interleukin-6 (IL-6) and tumor necrosis factor α (TNFα).