Gene Identification and Characterisation for Blinding
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Table S1 the Four Gene Sets Derived from Gene Expression Profiles of Escs and Differentiated Cells
Table S1 The four gene sets derived from gene expression profiles of ESCs and differentiated cells Uniform High Uniform Low ES Up ES Down EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol EntrezID GeneSymbol 269261 Rpl12 11354 Abpa 68239 Krt42 15132 Hbb-bh1 67891 Rpl4 11537 Cfd 26380 Esrrb 15126 Hba-x 55949 Eef1b2 11698 Ambn 73703 Dppa2 15111 Hand2 18148 Npm1 11730 Ang3 67374 Jam2 65255 Asb4 67427 Rps20 11731 Ang2 22702 Zfp42 17292 Mesp1 15481 Hspa8 11807 Apoa2 58865 Tdh 19737 Rgs5 100041686 LOC100041686 11814 Apoc3 26388 Ifi202b 225518 Prdm6 11983 Atpif1 11945 Atp4b 11614 Nr0b1 20378 Frzb 19241 Tmsb4x 12007 Azgp1 76815 Calcoco2 12767 Cxcr4 20116 Rps8 12044 Bcl2a1a 219132 D14Ertd668e 103889 Hoxb2 20103 Rps5 12047 Bcl2a1d 381411 Gm1967 17701 Msx1 14694 Gnb2l1 12049 Bcl2l10 20899 Stra8 23796 Aplnr 19941 Rpl26 12096 Bglap1 78625 1700061G19Rik 12627 Cfc1 12070 Ngfrap1 12097 Bglap2 21816 Tgm1 12622 Cer1 19989 Rpl7 12267 C3ar1 67405 Nts 21385 Tbx2 19896 Rpl10a 12279 C9 435337 EG435337 56720 Tdo2 20044 Rps14 12391 Cav3 545913 Zscan4d 16869 Lhx1 19175 Psmb6 12409 Cbr2 244448 Triml1 22253 Unc5c 22627 Ywhae 12477 Ctla4 69134 2200001I15Rik 14174 Fgf3 19951 Rpl32 12523 Cd84 66065 Hsd17b14 16542 Kdr 66152 1110020P15Rik 12524 Cd86 81879 Tcfcp2l1 15122 Hba-a1 66489 Rpl35 12640 Cga 17907 Mylpf 15414 Hoxb6 15519 Hsp90aa1 12642 Ch25h 26424 Nr5a2 210530 Leprel1 66483 Rpl36al 12655 Chi3l3 83560 Tex14 12338 Capn6 27370 Rps26 12796 Camp 17450 Morc1 20671 Sox17 66576 Uqcrh 12869 Cox8b 79455 Pdcl2 20613 Snai1 22154 Tubb5 12959 Cryba4 231821 Centa1 17897 -

C-Myc Regulates Cell Proliferation During Lens Development
c-myc Regulates Cell Proliferation during Lens Development Gabriel R. Cavalheiro., Gabriel E. Matos-Rodrigues., Anielle L. Gomes, Paulo M. G. Rodrigues, Rodrigo A. P. Martins* Programa de Biologia Celular e do Desenvolvimento, Instituto de Cieˆncias Biome´dicas, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Rio de Janeiro, Brazil Abstract Myc protooncogenes play important roles in the regulation of cell proliferation, growth, differentiation and survival during development. In various developing organs, c-myc has been shown to control the expression of cell cycle regulators and its misregulated expression is detected in many human tumors. Here, we show that c-myc gene (Myc) is highly expressed in developing mouse lens. Targeted deletion of c-myc gene from head surface ectoderm dramatically impaired ocular organogenesis, resulting in severe microphtalmia, defective anterior segment development, formation of a lens stalk and/or aphakia. In particular, lenses lacking c-myc presented thinner epithelial cell layer and growth impairment that was detectable soon after its inactivation. Defective development of c-myc-null lens was not caused by increased cell death of lens progenitor cells. Instead, c-myc loss reduced cell proliferation, what was associated with an ectopic expression of Prox1 and p27Kip1 proteins within epithelial cells. Interestingly, a sharp decrease in the expression of the forkhead box transcription factor Foxe3 was also observed following c-myc inactivation. These data represent the first description of the physiological roles played by a Myc family member in mouse lens development. Our findings support the conclusion that c- myc regulates the proliferation of lens epithelial cells in vivo and may, directly or indirectly, modulate the expression of classical cell cycle regulators in developing mouse lens. -

A Forkhead Gene, Foxe3, Is Essential for Lens Epithelial Proliferation and Closure of the Lens Vesicle
Downloaded from genesdev.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle Åsa Blixt,1 Margit Mahlapuu,1 Marjo Aitola,2 Markku Pelto-Huikko,2 Sven Enerba¨ck,1 and Peter Carlsson1,3 1Department of Molecular Biology, Go¨teborg University, The Lundberg Laboratory, Medicinaregatan 9C, Box 462, S-405 30 Go¨teborg, Sweden; 2Department of Developmental Biology, Medical School, University of Tampere, Tampere, Finland In the mouse mutant dysgenetic lens (dyl) the lens vesicle fails to separate from the ectoderm, causing a fusion between the lens and the cornea. Lack of a proliferating anterior lens epithelium leads to absence of secondary lens fibers and a dysplastic, cataractic lens. We report the cloning of a gene, FoxE3, encoding a forkhead/winged helix transcription factor, which is expressed in the developing lens from the start of lens placode induction and becomes restricted to the anterior proliferating cells when lens fiber differentiation begins. We show that FoxE3 is colocalized with dyl in the mouse genome, that dyl mice have mutations in the part of FoxE3 encoding the DNA-binding domain, and that these mutations cosegregate with the dyl phenotype. During embryonic development, the primordial lens epithelium is formed in an apparently normal way in dyl mutants. However, instead of the proliferation characteristic of a normal lens epithelium, the posterior of these cells fail to divide and show signs of premature differentiation, whereas the most anterior cells are eliminated by apoptosis. This implies that FoxE3 is essential for closure of the lens vesicle and is a factor that promotes survival and proliferation, while preventing differentiation, in the lens epithelium. -

Genome-Wide DNA Methylation Analysis of KRAS Mutant Cell Lines Ben Yi Tew1,5, Joel K
www.nature.com/scientificreports OPEN Genome-wide DNA methylation analysis of KRAS mutant cell lines Ben Yi Tew1,5, Joel K. Durand2,5, Kirsten L. Bryant2, Tikvah K. Hayes2, Sen Peng3, Nhan L. Tran4, Gerald C. Gooden1, David N. Buckley1, Channing J. Der2, Albert S. Baldwin2 ✉ & Bodour Salhia1 ✉ Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 diferentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in diferentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream efector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer. Activating KRAS mutations can be found in nearly 25 percent of all cancers1. -

Whole Exome Sequencing in Families at High Risk for Hodgkin Lymphoma: Identification of a Predisposing Mutation in the KDR Gene
Hodgkin Lymphoma SUPPLEMENTARY APPENDIX Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene Melissa Rotunno, 1 Mary L. McMaster, 1 Joseph Boland, 2 Sara Bass, 2 Xijun Zhang, 2 Laurie Burdett, 2 Belynda Hicks, 2 Sarangan Ravichandran, 3 Brian T. Luke, 3 Meredith Yeager, 2 Laura Fontaine, 4 Paula L. Hyland, 1 Alisa M. Goldstein, 1 NCI DCEG Cancer Sequencing Working Group, NCI DCEG Cancer Genomics Research Laboratory, Stephen J. Chanock, 5 Neil E. Caporaso, 1 Margaret A. Tucker, 6 and Lynn R. Goldin 1 1Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 2Cancer Genomics Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 3Ad - vanced Biomedical Computing Center, Leidos Biomedical Research Inc.; Frederick National Laboratory for Cancer Research, Frederick, MD; 4Westat, Inc., Rockville MD; 5Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; and 6Human Genetics Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA ©2016 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2015.135475 Received: August 19, 2015. Accepted: January 7, 2016. Pre-published: June 13, 2016. Correspondence: [email protected] Supplemental Author Information: NCI DCEG Cancer Sequencing Working Group: Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Anand Pathak, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, Guoqin Yu NCI DCEG Cancer Genomics Research Laboratory: Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. -

WO 2015/048577 A2 April 2015 (02.04.2015) W P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/048577 A2 April 2015 (02.04.2015) W P O P C T (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A61K 48/00 (2006.01) kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (21) International Application Number: BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, PCT/US20 14/057905 DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (22) International Filing Date: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, 26 September 2014 (26.09.2014) KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, (25) Filing Language: English PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, (26) Publication Language: English SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (30) Priority Data: 61/883,925 27 September 2013 (27.09.2013) US (84) Designated States (unless otherwise indicated, for every 61/898,043 31 October 2013 (3 1. 10.2013) US kind of regional protection available): ARIPO (BW, GH, GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, (71) Applicant: EDITAS MEDICINE, INC. -

Robles JTO Supplemental Digital Content 1
Supplementary Materials An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma based on mRNA, microRNA and DNA Methylation Biomarkers Ana I. Robles1, Eri Arai2, Ewy A. Mathé1, Hirokazu Okayama1, Aaron Schetter1, Derek Brown1, David Petersen3, Elise D. Bowman1, Rintaro Noro1, Judith A. Welsh1, Daniel C. Edelman3, Holly S. Stevenson3, Yonghong Wang3, Naoto Tsuchiya4, Takashi Kohno4, Vidar Skaug5, Steen Mollerup5, Aage Haugen5, Paul S. Meltzer3, Jun Yokota6, Yae Kanai2 and Curtis C. Harris1 Affiliations: 1Laboratory of Human Carcinogenesis, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 2Division of Molecular Pathology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 3Genetics Branch, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 4Division of Genome Biology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 5Department of Chemical and Biological Working Environment, National Institute of Occupational Health, NO-0033 Oslo, Norway. 6Genomics and Epigenomics of Cancer Prediction Program, Institute of Predictive and Personalized Medicine of Cancer (IMPPC), 08916 Badalona (Barcelona), Spain. List of Supplementary Materials Supplementary Materials and Methods Fig. S1. Hierarchical clustering of based on CpG sites differentially-methylated in Stage I ADC compared to non-tumor adjacent tissues. Fig. S2. Confirmatory pyrosequencing analysis of DNA methylation at the HOXA9 locus in Stage I ADC from a subset of the NCI microarray cohort. 1 Fig. S3. Methylation Beta-values for HOXA9 probe cg26521404 in Stage I ADC samples from Japan. Fig. S4. Kaplan-Meier analysis of HOXA9 promoter methylation in a published cohort of Stage I lung ADC (J Clin Oncol 2013;31(32):4140-7). Fig. S5. Kaplan-Meier analysis of a combined prognostic biomarker in Stage I lung ADC. -

Foxe3 Is Required for Morphogenesis and Differentiation of the Anterior Segment of the Eye and Is Sensitive to Pax6 Gene Dosage
Developmental Biology 302 (2007) 218–229 www.elsevier.com/locate/ydbio Foxe3 is required for morphogenesis and differentiation of the anterior segment of the eye and is sensitive to Pax6 gene dosage ⁎ Åsa Blixt a, Henrik Landgren a, Bengt R. Johansson b, Peter Carlsson a, a Dept. of Cell and Molecular Biology, Göteborg University, Box 462, 405 30 Göteborg, Sweden b The Electron Microscopy Unit, Dept. of Anatomy and Cell Biology, Göteborg University, Göteborg, Sweden Received for publication 12 June 2006; revised 8 September 2006; accepted 11 September 2006 Available online 16 September 2006 Abstract The dysgenetic lens (dyl) mouse mutant has mutations in Foxe3, which inactivate DNA binding by the encoded forkhead transcription factor. Here we confirm, by targeted inactivation, that Foxe3 mutations are responsible for the dyl phenotype, which include loss of lens epithelium; a small, cataractic lens; and failure of the lens to detach from the surface ectoderm. In contrast to a recent report of targeted Foxe3, we found no phenotypic difference between dyl and Foxe3−/− mutants when congenic strains were compared, and thus nothing that argues against Foxe3dyl being a null allele. In addition to the lens, most tissues of the anterior segment–iris, cornea, ciliary body and trabecular meshwork–are malformed or show differentiation defects. Many of these abnormalities, such as irido-corneal and irido-lenticular adherences, are present in a less severe form in mice heterozygous for the Foxe3 mutation, in spite of these having an intact lens epithelium. Early Foxe3 expression is highly sensitive to a halved Pax6 gene dosage and there is a striking phenotypic similarity between Pax6 and Foxe3 mutants. -
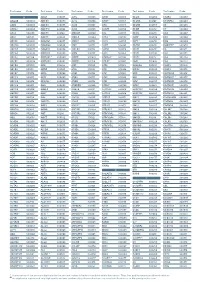
Aagab S00002 Aars S00003 Aars2 S00004 Aass S02483
Test name Code Test name Code Test name Code Test name Code Test name Code Test name Code A ADAR S00053 ALPL S00105 ARSB S00153 BCL10 S02266 C5AR2 S00263 AAGAB S00002 ADCK3 S00054 ALS2 S00106 ARSE * S00154 BCL11A S02167 C5ORF42 S00264 AARS S00003 ADCK4 S00055 ALX3 S00107 ARX S00155 BCL11B S02358 C6 S00265 AARS2 S00004 ADCY10 S02094 ALX4 S00108 ASAH1 S00156 BCOR S00212 C7 S00266 AASS S02483 ADCY3 S02184 AMACR S00109 ASL S00157 BCS1L S00213 C8A S00267 ABAT S02191 ADCY5 S02226 AMELX S02289 ASNS * S02508 BDNF S02509 C8B S00268 ABCA1 S00005 ADGRG1 S00057 AMER1 S00110 ASPA S00158 BDP1 * S00214 C8G S00269 ABCA12 S00006 ADGRG6 S02548 AMH S00111 ASPH S02425 BEAN1 S00215 C8ORF37 S00270 ABCA3 S00007 ADGRV1 S00058 AMHR2 S00112 ASPM S00159 BEST1 S00216 C9 S00271 ABCA4 S00008 ADIPOQ S00059 AMN S00113 ASS1 S00160 BFSP1 S02280 CA2 S00272 ABCA7 S02106 ADIPOR1 * S00060 AMPD1 S02670 ATAD3A * S02196 BFSP2 S00217 CA4 S02303 ABCB11 S00009 ADIPOR2 S00061 AMPD2 S02128 ATCAY S00162 BGN S02633 CA8 S00273 ABCB4 S00010 ADK S02595 AMT S00114 ATF6 S00163 BHLHA9 S00218 CABP2 S00274 ABCB6 S00011 ADNP S02320 ANG S00115 ATIC S02458 BICD2 S00220 CABP4 S00275 ABCB7 S00012 ADSL S00062 ANK1 S00116 ATL1 S00164 BIN1 S00221 CACNA1A S00276 ABCC2 S00013 AFF2 S00063 ANK2 S00117 ATL3 S00165 BLK S00222 CACNA1C * S00277 ABCC6 * S00014 AFG3L2 * S00064 ANKH S00118 ATM S00166 BLM S00223 CACNA1D S00278 ABCC8 S00015 AGA S00065 ANKRD11 * S02140 ATOH7 S02390 BLNK S02281 CACNA1F S00279 ABCC9 S00016 AGBL5 S02452 ANKS6 S00121 ATP13A2 S00168 BLOC1S3 S00224 CACNA1H S00280 ABCD1 * S00017 AGK * -

(12) Patent Application Publication (10) Pub. No.: US 2009/0269772 A1 Califano Et Al
US 20090269772A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0269772 A1 Califano et al. (43) Pub. Date: Oct. 29, 2009 (54) SYSTEMS AND METHODS FOR Publication Classification IDENTIFYING COMBINATIONS OF (51) Int. Cl. COMPOUNDS OF THERAPEUTIC INTEREST CI2O I/68 (2006.01) CI2O 1/02 (2006.01) (76) Inventors: Andrea Califano, New York, NY G06N 5/02 (2006.01) (US); Riccardo Dalla-Favera, New (52) U.S. Cl. ........... 435/6: 435/29: 706/54; 707/E17.014 York, NY (US); Owen A. (57) ABSTRACT O'Connor, New York, NY (US) Systems, methods, and apparatus for searching for a combi nation of compounds of therapeutic interest are provided. Correspondence Address: Cell-based assays are performed, each cell-based assay JONES DAY exposing a different sample of cells to a different compound 222 EAST 41ST ST in a plurality of compounds. From the cell-based assays, a NEW YORK, NY 10017 (US) Subset of the tested compounds is selected. For each respec tive compound in the Subset, a molecular abundance profile from cells exposed to the respective compound is measured. (21) Appl. No.: 12/432,579 Targets of transcription factors and post-translational modu lators of transcription factor activity are inferred from the (22) Filed: Apr. 29, 2009 molecular abundance profile data using information theoretic measures. This data is used to construct an interaction net Related U.S. Application Data work. Variances in edges in the interaction network are used to determine the drug activity profile of compounds in the (60) Provisional application No. 61/048.875, filed on Apr. -

Tracing Human Stem Cell Lineage During Development Using DNA Methylation
Downloaded from genome.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Research Tracing human stem cell lineage during development using DNA methylation Lucas A. Salas,1,8 John K. Wiencke,2,8 Devin C. Koestler,3 Ze Zhang,4,5 Brock C. Christensen,1,6,7,9 and Karl T. Kelsey4,5,9 1Department of Epidemiology, Geisel School of Medicine, Dartmouth College, Lebanon, New Hampshire 03756, USA; 2Department of Neurological Surgery, Institute for Human Genetics, University of California San Francisco, San Francisco, California 94158, USA; 3Department of Biostatistics, University of Kansas Medical Center, Kansas City, Kansas 66160, USA; 4Department of Epidemiology, 5Department of Pathology and Laboratory Medicine, Brown University, Providence, Rhode Island 02912, USA; 6Department of Molecular and Systems Biology, 7Department of Community and Family Medicine, Geisel School of Medicine, Dartmouth College, Lebanon, New Hampshire 03756, USA Stem cell maturation is a fundamental, yet poorly understood aspect of human development. We devised a DNA methyl- ation signature deeply reminiscent of embryonic stem cells (a fetal cell origin signature, FCO) to interrogate the evolving character of multiple human tissues. The cell fraction displaying this FCO signature was highly dependent upon develop- mental stage (fetal versus adult), and in leukocytes, it described a dynamic transition during the first 5 yr of life. Significant individual variation in the FCO signature of leukocytes was evident at birth, in childhood, and throughout adult life. The genes characterizing the signature included transcription factors and proteins intimately involved in embryonic develop- ment. We defined and applied a DNA methylation signature common among human fetal hematopoietic progenitor cells and have shown that this signature traces the lineage of cells and informs the study of stem cell heterogeneity in humans under homeostatic conditions. -

(12) Patent Application Publication (10) Pub. No.: US 2002/0177218 A1 Fang Et Al
US 2002O177218A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2002/0177218 A1 Fang et al. (43) Pub. Date: Nov. 28, 2002 (54) METHODS OF DETECTING MULTIPLE DNA Publication Classification BINDING PROTEIN AND DNA INTERACTIONS IN A SAMPLE, AND (51) Int. Cl." ....................................................... C12N 1/20 DEVICES, SYSTEMS AND KITS FOR (52) U.S. Cl. .......................................................... 435/252.3 PRACTICING THE SAME (57) ABSTRACT (76) Inventors: Yu Fang, Fremont, CA (US); Methods for detecting the presence of at least one, usually Xiao-Yang Wang, Mountain View, CA a plurality of, DNA binding proteins, e.g., transcription (US); Pierre Turpin, San Francisco, factors, in a Sample, both qualitatively and quantitatively, are CA (US) provided. In the Subject methods, a Substrate having one or Correspondence Address: more DNA probes immobilized on a surface thereof, one for each DNA binding protein of interest, is contacted with a BOZICEVIC, FIELD & FRANCIS LLP Sample under conditions Sufficient for binding complexes of 200 MIDDLEFIELD RID the probes and their respective DNA binding proteins to be SUTE 200 produced. The Sample may be purified with respect to one or MENLO PARK, CA 94025 (US) more DNA binding proteins or be a cellular/nuclear extract. (21) Appl. No.: 10/113,877 Resultant binding complexes on the Surface of the Substrate are then detected and related to the presence of the DNA (22) Filed: Mar. 29, 2002 binding protein-DAN interations of interest in the Sample. Also provided are devices and Systems for use in practicing Related U.S. Application Data the subject methods. The subject methods find use in a variety of different applications, e.g., detecting the presence (60) Provisional application No.