Population Structure Analysis of the Border Collie Dog Breed in Hungary
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Beauceron
The Beauceron The Beauceron, also known as Berger de Beauce and Bas Rouge, is the largest of the French sheepdogs and was developed solely in France with no foreign crosses. The Beauceron is closely related to the longhaired Briard or Berger de Brie. The first mention of a dog which matches the Beauceron’s description is found in a manuscript dated 1587. In 1809 Abbé Rozier wrote an article on French herding dogs. It was he who first described the differences in type and used the terms Berger de la Brie for long coated dogs and Berger de la Beauce for short coated dogs. The name Beauceron was used for the first time by Pierre Megnin in his 1888 book on war dogs and the first Berger de Beauce was registered with the Societe Central Canine in September 1893. The French Club Les Amis du Beauceron (CAB), was founded in 1922 by Pierre Megnin and he together with Emmanuel Boulet developed the original breed standard for the Beauceron. The CAB has since guided the development of the breed in its native France, always keeping a watchful eye on the preservation of the breed’s herding and working ability. During the early part of the 19th century large flocks of sheep were common and the Beauceron was indispensable for the shepherds of France; two dogs were sufficient to tend to flocks of 200 to 300 head of sheep. Sheep production experienced a sharp decline during the later half of the 19th century and by the second half of the 20th century was only a phantom of its past. -

Frequently Asked Questions
APPLYING SHAMPOO & CONDITIONER For best results apply the shampoo & conditioner in the following order: 1. belly & Determine the appropriate shampoo & conditioner for your breed. chest 2. legs and feet 3. neck & face 4. back SHORT COATS need LEMON, MEDIUM COATS need BANANA and bottom. Let soak for 5 full minutes. These LONG COATS need GREEN APPLE. are the sebaceous areas where most dirt and toxins enter the pet’s system. SHAMPOO: Dilute 1 part shampoo to 3 parts water in a clean container. Wet the coat & apply shampoo. Soak for 5 full minutes. Rinse. CONDITIONER: Dilute 1 part conditioner to 3 parts water in a clean container. Apply conditioner to coat. Soak for 5 full minutes. Rinse. Don’t see your breed? Please visit Please visit our info site http://isbusa.com/ instructions/akc-breeds-by-coat-type/ Frequently Asked Questions: SHORT COATS MEDIUM COATS LONG COATS Why are there different products for different American Foxhound Bernese Mountain Dog Afghan Hound breeds? All pet coats need oils, minerals and Staffordshire Terrier Brussels Griffon Bearded Collie collagen to thrive. However short coats need Basenji Cairn Terrier Bedlington Terrier more oils, medium coats need more minerals Basset Hound Cavalier King Charles Bichon Frise and long coats need more collagen. These Beagle Chesapeake Bay Retriever Bolonese products are created to provide the right Beauceron Chow Chow Chinese Crested balance of each for all coat types. Bloodhound Cocker Spaniel Coton De Tulear Why do I have to bathe per your instructions? Boston Terrier German Shepherd Havanese Since pets absorb dirt & toxins from the ground Boxer Golden Retriever Japanese Chin up into their sebaceous areas (tummy, feet, Great Dane Irish Wolfhound Lhasa Apso legs and other areas with less hair) it is Bull Terrier Keeshond Longhaired Dachshund important that the products are applied there Chihuahua Labrador Retriever Maltese first & allowed to soak for 5 min. -

Dog Breeds of the World
Dog Breeds of the World Get your own copy of this book Visit: www.plexidors.com Call: 800-283-8045 Written by: Maria Sadowski PlexiDor Performance Pet Doors 4523 30th St West #E502 Bradenton, FL 34207 http://www.plexidors.com Dog Breeds of the World is written by Maria Sadowski Copyright @2015 by PlexiDor Performance Pet Doors Published in the United States of America August 2015 All rights reserved. No portion of this book may be reproduced or transmitted in any form or by any electronic or mechanical means, including photocopying, recording, or by any information retrieval and storage system without permission from PlexiDor Performance Pet Doors. Stock images from canstockphoto.com, istockphoto.com, and dreamstime.com Dog Breeds of the World It isn’t possible to put an exact number on the Does breed matter? dog breeds of the world, because many varieties can be recognized by one breed registration The breed matters to a certain extent. Many group but not by another. The World Canine people believe that dog breeds mostly have an Organization is the largest internationally impact on the outside of the dog, but through the accepted registry of dog breeds, and they have ages breeds have been created based on wanted more than 340 breeds. behaviors such as hunting and herding. Dog breeds aren’t scientifical classifications; they’re It is important to pick a dog that fits the family’s groupings based on similar characteristics of lifestyle. If you want a dog with a special look but appearance and behavior. Some breeds have the breed characterics seem difficult to handle you existed for thousands of years, and others are fairly might want to look for a mixed breed dog. -

The History of the BERGER PICARD
The History of the BERGER PICARD By Betsy Richards hought to be the oldest upright ears, resembling in many ways a Beauce, or Beaceron). The mid-length of the French Sheep- Berger Picard of today. coat was ignored for some time, but final- dogs, the Berger Picard Of course it is not certain that the ly recognized as the Berger de Picardie was brought to northern Berger Picard originates strictly from the (or Picard). France and the Pas de Picardie region of France; it is possible, Although the Berger Picard made an Calais during the second even probable, that they were widespread appearance at the first French dog show TCeltic invasion of Gaul around 400 BC. as harsh-coated sheep and cattle dogs in 1863 and were judged in the same class Sheepdogs resembling Berger Picards were typical throughout northwestern as the Beaucerons and Briards, the breed's have been depicted for centuries in tapes- Europe. Some experts insist that this breed rustic appearance did not result in popu- tries, engravings and woodcuts. is related to the more well-known Briard larity as a show dog. In 1898 there was One renowned painting in the Bergerie and Beauceron, while others believe it proof that the Picard was a recognizable Nationale at Rambouillet (the National shares a common origin with Dutch and breed but it was tricolor, piebald with red Sheepfold of France) dating to the start Belgian Shepherds. brown markings. of the 19th century, shows the 1st Master Around the mid-19th century, dogs Picards would continue to be shown Shepherd, Clément Delorme, in the com- used for herding were initially classified and participate in herding trials but strug- pany of a medium-sized, strong-boned dog as one of two types: long hair (Berger de gled for recognition. -

Ranked by Temperament
Comparing Temperament and Breed temperament was determined using the American 114 DOG BREEDS Popularity in Dog Breeds in Temperament Test Society's (ATTS) cumulative test RANKED BY TEMPERAMENT the United States result data since 1977, and breed popularity was determined using the American Kennel Club's (AKC) 2018 ranking based on total breed registrations. Number Tested <201 201-400 401-600 601-800 801-1000 >1000 American Kennel Club 50% 60% 70% 80% 90% 1. Labrador 100% Popularity Passed 2. German Retriever Passed Shepherd 3. Mixed Breed 7. Beagle Dog 4. Golden Retriever More Popular 8. Poodle 11. Rottweiler 5. French Bulldog 6. Bulldog (Miniature)10. Poodle (Toy) 15. Dachshund (all varieties) 9. Poodle (Standard) 17. Siberian 16. Pembroke 13. Yorkshire 14. Boxer 18. Australian Terrier Husky Welsh Corgi Shepherd More Popular 12. German Shorthaired 21. Cavalier King Pointer Charles Spaniel 29. English 28. Brittany 20. Doberman Spaniel 22. Miniature Pinscher 19. Great Dane Springer Spaniel 24. Boston 27. Shetland Schnauzer Terrier Sheepdog NOTE: We excluded breeds that had fewer 25. Bernese 30. Pug Mountain Dog 33. English than 30 individual dogs tested. 23. Shih Tzu 38. Weimaraner 32. Cocker 35. Cane Corso Cocker Spaniel Spaniel 26. Pomeranian 31. Mastiff 36. Chihuahua 34. Vizsla 40. Basset Hound 37. Border Collie 41. Newfoundland 46. Bichon 39. Collie Frise 42. Rhodesian 44. Belgian 47. Akita Ridgeback Malinois 49. Bloodhound 48. Saint Bernard 45. Chesapeake 51. Bullmastiff Bay Retriever 43. West Highland White Terrier 50. Portuguese 54. Australian Water Dog Cattle Dog 56. Scottish 53. Papillon Terrier 52. Soft Coated 55. Dalmatian Wheaten Terrier 57. -

Agtsec Running Groups
Group A - 142 competitors Champ: 10 - 8, 14 - 5, 16 - 12, 20 - 15, 22 - 18, 24 - 12 Perf: P8 - 9, P12 - 6, P14 - 8, P16 - 10, P20 - 9 Vet: V4 - 4, V8 - 5, V12 - 17, V16 - 4 Addison, Michelle DeChance, Annie Lady (24) German Shepherd Dog Pink Floyd (V12) Token Blonde Spencer Davis (P20) Rico Suave Dog Alfonso, Annette Chapter (22) Border Collie Erspamer, Mia Legend (V12) Border Collie Jackson (P20) Labrador Retriever Valid (P20) Border Collie Anderson, Cliff Zoe (20) Wheatable Ferguson, Kelley Winnie (V12) Border Collie Joose (16) Border Collie Anderson, Crystal Floyd, Cindy Razzi (P14) English Springer Spaniel Thor (16) Poodle (Miniature) Andrews, Lisa Friedl, Gwyneth Shibumi (24) Border Collie Amigo (24) Border Collie Aubois, Sara Gant, Shane Ridley (P20) Border Collie Atom (P20) Border Collie Sweets (P12) Shetland Sheepdog Barton, Kim Logan (V 4) Shih Tzu Garcia, Allison EPI (20) All-American Sizzle (V 8) Shetland Sheepdog Better Cheddar (14) All-American Bekaert, Susan Ringer (P16) Border Collie ABBA (V12) Border Collie Motown (16) Shetland Sheepdog Garvey, Sarah Poppy (24) All-American Bennett, Alicia Excalibur (V16) Border Collie Gerhard, Jeremy Bleu (10) Papillon Maverick (10) Pembroke Welsh Corgi Pixie Pig (20) Border Collie Tease (20) Border Collie Ruckus (22) Australian Shepherd Benson, Helen Shadow (16) Shetland Sheepdog Grace, Kathy Blanche (16) Standard Schnauzer Bowman, Tom Casey (P14) All-American Hanson, Morgan Probability (P16) Border Collie Brown, Kat #Winning (22) Border Collie Nemo (P14) All-American Elite (20) Border -
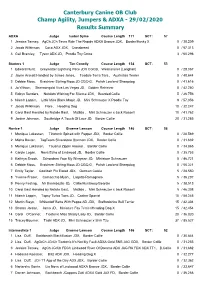
29/02/2020 Results Summary
Canterbury Canine OB Club Champ Agility, Jumpers & ADXA - 29/02/2020 Results Summary ADXA Judge Isobel Syme Course Length 171 SCT: 57 1 Jessica Tansey, AgCh JCh Tevra Ride The Rapids ADXA Bronze JDX, Border/Husky X 0 / 38.209 2 Jacob Wilkinson, Coco ADX JDX, Crossbreed 0 / 57.313 3 Gail Bramley, Tyson ADX JD, Poodle Toy Cross 3 / 60.296 Starters 1 Judge Tim Conolly Course Length 134 SCT: 53 1 Edward Hunt, Greymatter Lightning Pace JDX CGCB, Weimaraner (Longhair) 0 / 28.067 2 Jayne Arscott Handled by James Jones, Tasdale Terra Tara , Australian Terrier 0 / 40.644 3 Debbie Moss, Braidriver Stirling Moss JD CGC-G, Polish Lowland Sheepdog 0 / 41.616 4 Jo Wilson, Shannongold Viva Las Vegas JD, Golden Retriever 0 / 42.250 5 Robyn Sanders, Notslots Wishing For Silence JDX, Bearded Collie 5 / 36.756 6 Niamh Lappin, Little Miss Black Magic JD, Mini Schnauzer X Poodle Toy 9 / 57.356 7 Jacob Wilkinson, Flare , Heading Dog 10 / 32.247 8 Carol East Handled by Natalie East, Matilda , Mini Schnauzer x Jack Russell 10 / 41.762 9 Janine Johnson, Southridge A Touch Of Lace JD, Border Collie 20 / 31.285 Novice 1 Judge Graeme Lawson Course Length 146 SCT: 58 1 Monique Lukassen, Ticotonic Spiced with Pepper JDX, Border Collie 0 / 28.569 2 Misha Baxter, TagTeam Silverstone Starman JDX, Border Collie 0 / 31.632 3 Monique Lukassen, Tauanui Zippin Around , Border Collie 0 / 34.865 4 Carole Logan, Nova Echo of Lindwood JD, Border Collie 0 / 35.753 5 Kathryn Snook, Schondara Your My Wingman JD, Miniature Schnauzer 0 / 46.721 6 Debbie Moss, Braidriver Stirling Moss -

HERDING BREEDS ELIGIBLE to COMPETE in ASCA STOCKDOG TRIALS Ref: the Atlas of Dog Breeds of the World - Bonnie Wilcox, DVM & Chris Walkowic
ASCA STOCKDOG PROGRAM RULES – JUNE 2014 APPENDIX 6: HERDING BREEDS ELIGIBLE TO COMPETE IN ASCA STOCKDOG TRIALS Ref: The Atlas of Dog Breeds of the World - Bonnie Wilcox, DVM & Chris Walkowic Breed Country of Origin Breed Country of Origin Australian Cattle Dog Australia Greater Swiss Mountain Dog Switzerland Australian Kelpie Australia Hairy Mouth Heeler USA Australian Shepherd USA Hovawart Germany Belgian Laekenois Belgium Iceland Dog Iceland Belgian Malinois Belgium Kerry Blue Terrier Ireland Belgian Sheep Dog Lancashire Heeler Great Britain (Groenendael) Belgium Lapinporokoira Belgian Tervuren Belgium (Lapponian Herder) Finland Bouviers Des Flandres Belgium Malinois Belgian Bergamasco Italy McNab USA Bernese Mtn Dog Switzerland Miniature Australian Shepherd USA Beauceron France Mudi Hungary Briard France North American Shepherd USA Bearded Collie Great Britain Norwegian Buhund Norway Border Collie Great Britain Old English Sheep Dog Great Britain Blue Lacy USA Picardy Shepherd France Catahoula Leopard Dog USA Polish Owczarek Nizinny Poland Canaan Dog Israel Puli Hungary Cao de Serra de Aires Pumi Hungary (Portuguese Sheep Dog) Portugal Pyrenean Shepherd France Croatian Sheep Dog Croatia Rottweiler Germany Cataian Sheep Dog Spain Samoyed Scandinavia Collie Great Britain Schapendoes Corgi (Dutch Sheepdog) Netherlands Cardigan Welsh Great Britain Shetland Sheep Dog Great Britain Pembroke Welsh Great Britain Soft Coated Wheaten Terrier Ireland Dutch Shepherd: Netherlands Spanish Water Dog Spain English Shepherd USA Swedish Lapphund Sweden Entlebucher Mountain Dog Switzerland Tibetan Terrier China Finnish Lapphund Finland Vasgotaspets Sweden German Shepherd Dog Germany White Shepherd USA German Coolie/Koolie Australia 74 . -

Commencing Implementation of a Genetic Evaluation System for Livestock Working Dogs by C
Commencing implementation of a genetic evaluation system for livestock working dogs by C. M. Wade, D. van Rooy, E. R. Arnott, J. B. Early and P. D. McGreevy June 2021 Commencing implementation of a genetic evaluation system for livestock working dogs by C. M. Wade, D. van Rooy, E. R. Arnott, J. B. Early and P. D. McGreevy June 2021 i © 2021 AgriFutures Australia All rights reserved. ISBN 978-1-76053-135-5 ISSN 1440-6845 Commencing implementation of a genetic evaluation for livestock working dogs Publication No. 20-117 Project No: PRJ-010413 The information contained in this publication is intended for general use to assist public knowledge and discussion and to help improve the development of sustainable regions. You must not rely on any information contained in this publication without taking specialist advice relevant to your particular circumstances. While reasonable care has been taken in preparing this publication to ensure that information is true and correct, the Commonwealth of Australia gives no assurance as to the accuracy of any information in this publication. The Commonwealth of Australia, AgriFutures Australia, the authors or contributors expressly disclaim, to the maximum extent permitted by law, all responsibility and liability to any person, arising directly or indirectly from any act or omission, or for any consequences of any such act or omission, made in reliance on the contents of this publication, whether or not caused by any negligence on the part of the Commonwealth of Australia, AgriFutures Australia, the authors or contributors. The Commonwealth of Australia does not necessarily endorse the views in this publication. -

Downloading Or Purchasing Online At
Valuable behavioural phenotypes in Australian farm dogs PD McGreevy, CM Wade, ER Arnott and JB Early Front cover photo credit: Lorraine Williams October 2015 RIRDC Publication No 15/081 RIRDC Project No: PRJ-007806 © 2015 Rural Industries Research and Development Corporation. All rights reserved. ISBN 978-1-74254-827-2 ISSN 1440-6845 Valuable behavioural phenotypes in Australian farm dogs Publication No. 15/081 Project No. PRJ-007806 The information contained in this publication is intended for general use to assist public knowledge and discussion and to help improve the development of sustainable regions. You must not rely on any information contained in this publication without taking specialist advice relevant to your particular circumstances. While reasonable care has been taken in preparing this publication to ensure that information is true and correct, the Commonwealth of Australia gives no assurance as to the accuracy of any information in this publication. The Commonwealth of Australia, the Rural Industries Research and Development Corporation (RIRDC), the authors or contributors expressly disclaim, to the maximum extent permitted by law, all responsibility and liability to any person, arising directly or indirectly from any act or omission, or for any consequences of any such act or omission, made in reliance on the contents of this publication, whether or not caused by any negligence on the part of the Commonwealth of Australia, RIRDC, the authors or contributors. The Commonwealth of Australia does not necessarily endorse the views in this publication. This publication is copyright. Apart from any use as permitted under the Copyright Act 1968, all other rights are reserved. -

Sixth Akc Rally® National Championship
SIXTH AKC RALLY® NATIONAL CHAMPIONSHIP FRIDAY, MARCH 15, 2019 CENTRAL PARK HALL AT EXPO SQUARE 4145 EAST 21ST STREET – TULSA, OK 74114 TWENTY FIFTH AKC NATIONAL OBEDIENCE CHAMPIONSHIP SATURDAY & SUNDAY MARCH 16-17, 2019 AKC MISSION STATEMENT The American Kennel Club is dedicated to upholding the integrity of its Registry, promoting the sport of purebred dogs and breeding for type and function. Founded in 1884, the AKC® and its affiliated organizations advocate for the purebred dog as a family companion, advance canine health and well-being, work to protect the rights of all dog owners and promote responsible dog ownership. AKC OBJECTIVE Advance the study, breeding, exhibiting, running and maintenance of purebred dogs. AKC CORE VALUES We love purebred dogs. We are committed to advancing the sport of the purebred dog. We are dedicated to maintaining the integrity of our Registry. We protect the health and well-being of all dogs. We cherish dogs as companions. We are committed to the interests of dog owners. We uphold high standards for the administration and operation of the AKC. We recognize the critical importance of our clubs and volunteers. SIXTH AKC RALLY® NATIONAL CHAMPIONSHIP FRIDAY, MARCH 15, 2019 Sponsored in part by Eukanuba™ and J & J Dog Supplies TWENTY FIFTH AKC NATIONAL OBEDIENCE CHAMPIONSHIP SATURDAY & SUNDAY MARCH 16-17, 2019 Permission is granted by the American Kennel Club for the holding of this event under American Kennel Club rules and regulations. 1 Gina M. DiNardo, Secretary AKC BOARD OF DIRECTORS Ronald H. Menaker – Chairman Dr. Thomas M. Davies – Vice Chairman Class of 2019 Class of 2020 • Dr. -

Merle Phenotypes in Dogs – SILV SINE Insertions from Mc to Mh
bioRxiv preprint doi: https://doi.org/10.1101/328690; this version posted May 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Merle phenotypes in dogs – SILV SINE insertions from Mc to Mh Mary Langevin*, Helena Synkova**, Tereza Jancuskova***, Sona Pekova+*** *Cat´s Cradle Catahoulas, Oro Medonte, Ontario, Canada **European Association of Louisiana Catahoulas, K. Chocholy 16, 370 05 Ceske Budejovice, Czech Republic ***Vemodia a.s., Laboratory for veterinary molecular diagnostics, V Hurkach 3, Prague, Czech Republic +Corresponding author: [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/328690; this version posted May 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. ABSTRACT It has been recognized that the Merle coat pattern is not only a visually interesting feature, but it also exerts an important biological role, in terms of hearing and vision impairments. In 2006, the Merle (M) locus was mapped to the SILV gene with a SINE element in it, and the inserted retroelement was proven causative to the Merle phenotype. Mapping of the M locus was a genetic breakthrough and many breeders started implementing SILV SINE testing in their breeding programs. Unfortunately, the situation turned out complicated as genotypes of Merle tested individuals did not always correspond to expected phenotypes, sometimes with undesired health consequences in offspring.