Interplay Between NS3 Protease and Human La Protein Regulates
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

List of Life Members As on 20Th January 2021
LIST OF LIFE MEMBERS AS ON 20TH JANUARY 2021 10. Dr. SAURABH CHANDRA SAXENA(2154) ALIGARH S/O NAGESH CHANDRA SAXENA POST HARDNAGANJ 1. Dr. SAAD TAYYAB DIST ALIGARH 202 125 UP INTERDISCIPLINARY BIOTECHNOLOGY [email protected] UNIT, ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 11. Dr. SHAGUFTA MOIN (1261) [email protected] DEPT. OF BIOCHEMISTRY J. N. MEDICAL COLLEGE 2. Dr. HAMMAD AHMAD SHADAB G. G.(1454) ALIGARH MUSLIM UNIVERSITY 31 SECTOR OF GENETICS ALIGARH 202 002 DEPT. OF ZOOLOGY ALIGARH MUSLIM UNIVERSITY 12. SHAIK NISAR ALI (3769) ALIGARH 202 002 DEPT. OF BIOCHEMISTRY FACULTY OF LIFE SCIENCE 3. Dr. INDU SAXENA (1838) ALIGARH MUSLIM UNIVERSITY, ALIGARH 202 002 HIG 30, ADA COLONY [email protected] AVANTEKA PHASE I RAMGHAT ROAD, ALIGARH 202 001 13. DR. MAHAMMAD REHAN AJMAL KHAN (4157) 4/570, Z-5, NOOR MANZIL COMPOUND 4. Dr. (MRS) KHUSHTAR ANWAR SALMAN(3332) DIDHPUR, CIVIL LINES DEPT. OF BIOCHEMISTRY ALIGARH UP 202 002 JAWAHARLAL NEHRU MEDICAL COLLEGE [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 14. DR. HINA YOUNUS (4281) [email protected] INTERDISCIPLINARY BIOTECHNOLOGY UNIT ALIGARH MUSLIM UNIVERSITY 5. Dr. MOHAMMAD TABISH (2226) ALIGARH U.P. 202 002 DEPT. OF BIOCHEMISTRY [email protected] FACULTY OF LIFE SCIENCES ALIGARH MUSLIM UNIVERSITY 15. DR. IMTIYAZ YOUSUF (4355) ALIGARH 202 002 DEPT OF CHEMISTRY, [email protected] ALIGARH MUSLIM UNIVERSITY, ALIGARH, UP 202002 6. Dr. MOHAMMAD AFZAL (1101) [email protected] DEPT. OF ZOOLOGY [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 ALLAHABAD 7. Dr. RIAZ AHMAD(1754) SECTION OF GENETICS 16. -

Awardees of National Bioscience Award for Career Development
AWARDEES OF NATIONAL BIOSCIENCE AWARD FOR CAREER DEVELOPMENT Awardees for the year 2016 1. Dr. Mukesh Jain, Associate Professor, School of Computational and Integrative Sciences, Jawaharlal Nehru University, New Delhi-110067 2. Dr. Samir K. Maji, Associate Professor, Indian Institute of Technology, Powai, Mumbai- 400076 3. Dr. Anindita Ukil, Assistant Professor, Calcutta University, Kolkata 4. Dr. Arnab Mukhopadhyay, Staff Scientist V, National Institute of Immunology, Aruna Asaf Ali Marg, New Delhi- 110067 5. Dr. Rohit Srivastava, Professor, Indian Institute of Technology, Bombay, Mumbai- 400076 6. Dr. Pinaki Talukdar, Associate Professor, Indian Institute of Science Education and Research, Dr. Homi Bhabha Road, Pashan, Pune- 7. Dr. Rajnish Kumar Chaturvedi, Senior Scientist, CSIR- Indian Institute of Toxicology Research, Lucknow-226001 8. Dr. Jackson James, Scientist E-II, Neuro Stem Cell Biology Lab, Neurobiology Division, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala- 695014 Awardees for the year 2015 1. Dr. Sanjeev Das, Staff Scientist-V, National Institute of Immunology, New Delhi 2. Dr. Ganesh Nagaraju, Assistant Professor, Department of Biotechnology, Indian Institute of Science, Bangalore- 5600012. 3. Dr. Suvendra Nath Bhattacharya, Principal Scientist, CSIR- Indian Institute of Chemical Biology, Kolkata- 700032 4. Dr. Thulasiram H V, Principal Scientist, CSIR-National Chemical Laboratory, Pune- 411008. 5. Dr. Pawan Gupta, Principal Scientist, Institute of microbial Technology, Chandigarh- 160036. 6. Dr. Souvik Maiti, Principal Scientist, CSIR-Institute of Genomics and Integrative Biology, Delhi- 110025. 7. Dr. Pravindra Kumar, Associate Professor, Department of Biotechnology, IIT, Roorkee- 247667. 8. Dr. Anurag Agrawal, Principal Scientist, CSIR-Institute of Genomics and Integrative Biology, Delhi- 110025 9. Dr. Gridhar Kumar Pandey, Professor, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi- 110067 10. -

National Bioscience Awards for Career Development
AWARDEES OF NATIONAL BIOSCIENCE AWARDS FOR CAREER DEVELOPMENT Awardees for the year 2012 1. Dr. Kaustuv Sanyal, Associate Professor, Molecular Mycology Laboratory, Molecular Biology & Genetics Unit, Jawaharlal Nehru Centre for Advance Scientific Research, Jakkur P.O. Bangalore 560064 2. Dr Naval Kishore Vikram, Associate Professor, Department of Medicine, All India Institute of Medical Sciences (AIIMS), Ansari Nagar, New Delhi- 110029 3. Dr. Aditya Bhushan Pant, Senior Scientist & In-charge, In Vitro Toxicology Laboratory, Indian Institute of Toxicology Research, PO Box: 80, MG Marg, Lucknow 226001 (UP) India 4. Dr. Subrata Adak, Senior Scientist, Indian Institute of Chemical Biology; 4, Raja S.C. Mullick Road, Kolkata-700032 5. Dr. Durai Sundar, Assistant Professor, Dept of Biochemical Engineering & Biotechnology, Indian Institute of Technology (IIT) Delhi, Hauz Khas, New Delhi – 110016 6. Dr S Venkata Mohan, Senior Scientist, Bioengineering and Environmental Centre (BEEC) CSIR-Indian Institute of Chemical Technology, Hyderabad-500 607 7. Dr. Munia Ganguli, Scientist E-I, CSIR-Institute of Genomics & Integrative Biology, Mall Road,New Delhi 110 007 8. Dr. Asad U Khan, Associate Professor & Coordinator/Head of Biotechnology Department, A.M.U, Interdisciplinary Biotechnology Unit, A.M.U., Aligarh 202002 9. Dr. Sathees C. Raghavan, Assistant Professor, Department of Biochemistry, Indian Institute of Science, Bangalore 560 012 10. Dr. Vidita A. Vaidya, Associate Professor, Department of Biological Sciences, Tata Institute of Fundamental Research, 1, Homi Bhabha Road, Colaba, Mumbai - 400005 Awardees for the year 2011 1. Dr. M. M. Parida, Scientist-F, Joint Director, Division of Virology Defence R & D Establishment, DRDE, DRDO, Ministry of Defence, Jhansi Road, Gwalior- 474002 2. -
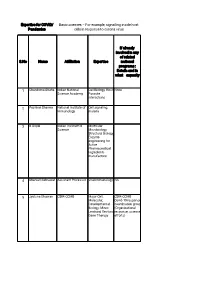
Basic Sciences – for Example, Signalling Inside Host Pandemics Cells in Response to Corona Virus
Expertise for COVID/ Basic sciences – For example, signalling inside host Pandemics cells in response to corona virus If already involved in any of related S.No Name Affiliation Expertise national programs : Details and in what capacity 1 Chandrima Shaha Indian National Cell Biology, Host None Science Academy Parasite interactions 2 Pushkar Sharma National Institute of Cell signaling, Immunology malaria 3 B Gopal Indian Institute of Molecular Science Microbiology, Structural Biology, Enzyme engineering for Active Pharmaceutical Ingredients manufacture 4 Sharvan Sehrawat Assistant Professor Viral immunology NA 5 Jyotsna Dhawan CSIR-CCMB Major-Cell, CSIR-CCMB Molecular, Covid-19 response Developmental coordination group Biology; Minor: (Organizational Lentiviral Vectors, response, science Gene Therapy efforts) 6 G. Baskaran The Institute of Theoretical Mathematical Physics. Sciences, Chennai Curious about 600 113 and biological IITMadras processes for decades. 7 Pradip Kumar Jamia Hamdard, NewMolecular NA Chakraborti Delhi 110062 Microbiology, Infectious disease, prokaryotic signalling 8 Nahid Ali CSIR Indian institute Parasitology, of chemical biology immunology 9 Raj Kumar Roy IISER Mohali Polymer and No Material Chemistry 10 Narinder K. Mehra All India Institute of COVID-19 and Medical Sciences immunity, Laboratory diagnosis 11 V. Ramanathan IIT(BHU) Varanasi Raman imaging Have been helping for disease the administrative diagnosis authorities of my city and my institute in making and distributing hand sanitizers. Till date have made around -
Inside This Issue
NEWS JNC Issue - 38 Jawaharlal Nehru Centre for Advanced Scientific Research May 2012 Jakkur P.O., Bangalore-64 (A Deemed to be University) Focus Dear Colleagues The Centre has been recording an impressive growth in research and academic programmes. The recognition that the faculty members have received is an evidence of such progressive endeavour. Prof. CNR Rao has been honoured with the Einstein Professorship of Chinese Academy of Sciences. It’s heartening to know that JNCASR is ranked one among the top 10 Indian institutions in quality proxies as per SCImago Institutions Ranking (SIR) World Report 2011 based on Scopus data. Let me congratulate all the faculty colleagues for this unique distinction. The new infrastructural facilities like CCMS, EOBU Lab, Nano Sciences, Seminar/Lecture Hall Complex are coming up and a new visiting students hostel is already ready and functional. M R S Rao President inside this issue... From the Editor’s Desk Fluid Dynamics Colloquia Short Term Appointments Discussion Meetings Intellectual Property Schools/Workshops/Symposia Awards and Recognitions Seminars International Centre for Materials Other Programmes Science Forthcoming Meetings Endowment Lectures Annual Faculty Meeting From The Editor’s Desk The Centre has recorded some significant progress in MoUs have been signed with RAK-CAM in December academic and extension activities. The current student 2011 and with DFRL in January 2012. strength is 268. 8 students had joined the Centre for the mid-year admissions of 2011 -12 during January "Chemistry Program" in celebration of IYC-2011 was 2012. organized at C N R Rao Hall of Science on December 7, 2011. -

Annual Report 2016-2017
CSIR-IICB CSIR-Indian Institute of Chemical Biology Annual Report 2016-17 #$%&' ! " ! #$%&' ( ! " ! #$%&' ( Courtesy : Dr. Suvendra Nath Bhattacharya Annual Report 2016-17 Jadavpur Campus Salt Lake Campus CSIR-Indian Institute of Chemical Biology 4, Raja S. C. Mullik Road, Jadavpur, Kolkata - 700 032, India Cancer Biology & Project Monitoring Inflammatory Disorder & Evaluation Infectious Diseases Art & & Immunology Photography Cell Biology Business & Physiology Development Group Molecular Genetics ISTAD Organic & Medicinal Chemistry IPM Cell Structural Biology & Bio-informatics P&I R & D P&I - PME & BDG Divisions Division Animal House Electrical Engineering Library & Documentation Maintenance CSIR Electrical Instrumentation IICB Central Architecture Facilities & Civil Engineering Kolkata Infrastructure Computer Maintenance (Civil) Human Resource Group Administration Director’s Finance & Secretariate Accounts General Stores & Administration Purchase Establishment Bill General Section Store R & C Cash Foreign Section Section Purchase Security & Receipts & Indigenous Maintenance Despatch Purchase Works & Hindi Cell Services Contents Director’s Report 005 Cell Biology & Physiology Division 009 Infectious Diseases & Immunology Division 027 Cancer Biology & Inflammatory Disorders Division 037 Organic & Medicinal Chemistry Division 059 Molecular Genetics Division 079 Structural Biology & Bioinformatics Division 085 Network Projects in 12th Five year Plan (2012-17) 105 Publication -

RESUME Dr. B. UMA REDDY Associate Professor PG Department
RESUME Dr. B. UMA REDDY Associate Professor PG Department of studies in Botany Vijayanagara Srikrishnadevaraya University Ballari- 583104 Email: [email protected] EDUCATION Senior Research Associate (SRA): Dept of Microbiology and Cell Biology, Indian Institute of Science, Bangalore. Aug 2010 to Jan DST- FAST-TRACK Young Scientist: Completed DST, GOI sponsored project 2019. (Rs. ~30 lakhs/-) approved under FAST-TRACK young Scientists Scheme, May 2012-15) at Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore. 2008-2009 PG diploma in Nanoscience & Technology, Gulbarga University, Gulbarga 1996-2001 Ph.D. Gulbarga University Gulbarga 1995-1996 M.Phil. (Phytochemistry and Pharmacology), Gulbarga University, Gulbarga 1993-1995 M. Sc. (Botany, spl. in medicinal plants), Gulbarga University, Gulbarga EMPLOYMENT From 27th May Associate Professor, PG Department of Studies in Botany, Vijayanagara 2019 onwards Srikrishnadevaraya University. Feb, 2019 to 24th Coordinator for Nanobiotechnology Training Program, Centre for May 2019. Nanoscience and Engineering, Indian Institute of Science. Guest Lecturer: Botany, Bioinformatics Dept, Gulbarga University, 2008-10 Gulbarga. Lecturer: Biotechnology Dept, National College [private], Bellary, Karnataka 2004-08 (2004-08). 2000-03 Lecturer: Smt. A.S.M. College for Women, Bellary (2000-03). Sponsored Projects under Supervision as Principal Investigator and Funds Received Project Title Sponsoring Agency Amount Reference “Screening of medicinal Science and Engineering Research 30 lakhs No.SR/FT/LS-01/2011 plants for antiiral Board DST, Govt of India properties against SERB-DST Fast Track Hepatitis C virus” Technology Bhavan Young Scientists New Meharauli Road scheme New Delhi 110016 1 AWARDS / HONORS / PRIZES Awarded DST Fast Track Young Scientists Project as a Principal Investigator at IISc Bangalore. -

Indian Institute of Science, Bangalore 560 012
IISc Profile 2012 Indian Institute of Science, Bangalore 560 012 CONTENTS Foreword v The Court viii The Council ix Administration x Division of Biological Sciences Department of Biochemistry 2 Department of Microbiology and Cell Biology 4 Department of Molecular Reproduction, Development and Genetics 6 Molecular Biophysics Unit 8 Centre for Ecological Sciences 10 Centre for Neuroscience 12 Central Animal Facility 14 Primate Research Laboratory 16 Division of Chemical Sciences Department of Inorganic and Physical Chemistry 18 Department of Organic Chemistry 20 Solid State and Structural Chemistry Unit 22 Materials Research Centre 24 NMR Research Centre 26 Division of Physical and Mathematical Sciences Department of Instrumentation and Applied Physics 28 Department of Mathematics 30 Department of Physics 32 Astronomy and Astrophysics Programme 34 Centre for High Energy Physics 36 Centre for Contemporary Studies 38 Centre for Cryogenic Technology 40 Division of Electrical Sciences Department of Computer Science and Automation 42 Department of Electrical Engineering 44 Department of Electrical Communication Engineering 46 Department of Electronic Systems Engineering 48 Division of Mechanical Sciences Department of Aerospace Engineering 50 Department of Chemical Engineering 56 Department of Mechanical Engineering 58 Department of Materials Engineering 62 Centre for Product Design and Manufacturing 64 iii iv Contents Division of Earth and Environmental Sciences Department of Civil Engineering 66 Department of Management Studies 70 Centre for -

LIST of LIFE MEMBERS AS on 1St SEPTEMBER 2020
LIST OF LIFE MEMBERS AS ON 1st SEPTEMBER 2020 10. Dr. SHAGUFTA MOIN (1261) ALIGARH DEPT. OF BIOCHEMISTRY J. N. MEDICAL COLLEGE 1. Dr. HAMMAD AHMAD SHADAB G. G.(1454) ALIGARH MUSLIM UNIVERSITY 31 SECTOR OF GENETICS ALIGARH 202 002 DEPT. OF ZOOLOGY ALIGARH MUSLIM UNIVERSITY 11. SHAIK NISAR ALI (3769) ALIGARH 202 002 DEPT. OF BIOCHEMISTRY FACULTY OF LIFE SCIENCE 2. Dr. INDU SAXENA (1838) ALIGARH MUSLIM UNIVERSITY HIG 30, ADA COLONY ALIGARH 202 002 AVANTEKA PHASE I [email protected] RAMGHAT ROAD, ALIGARH 202 001 12. DR. MAHAMMAD REHAN AJMAL KHAN (4157) 3. Dr. (MRS) KHUSHTAR ANWAR SALMAN(3332) 4/570, Z-5, NOOR MANZIL COMPOUND DEPT. OF BIOCHEMISTRY DIDHPUR, CIVIL LINES JAWAHARLAL NEHRU MEDICAL COLLEGE ALIGARH UP 202 002 ALIGARH MUSLIM UNIVERSITY [email protected] ALIGARH 202 002 [email protected] 13. DR. HINA YOUNUS (4281) INTERDISCIPLINARY BIOTECHNOLOGY UNIT 4. Dr. MOHAMMAD TABISH (2226) ALIGARH MUSLIM UNIVERSITY DEPT. OF BIOCHEMISTRY ALIGARH U.P. 202 002 FACULTY OF LIFE SCIENCES [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 [email protected] ALLAHABAD 5. Dr. MOHAMMAD AFZAL (1101) 14. Dr. B. SHARMA (872) DEPT. OF ZOOLOGY DEPARTMENT OF BIOCHEMISTRY ALIGARH MUSLIM UNIVERSITY UNIVERSITY OF ALLAHABAD ALIGARH 202 002 ALLAHABAD 211002 INDIA [email protected] 6. Dr. RIAZ AHMAD(1754) [email protected] SECTION OF GENETICS 9415715639 DEPT. OF ZOOLOGY ALIGARH MUSLIM UNIVERSITY 15. Dr. ABHAY KUMAR PANDEY (2054) ALIGARH 202 002 DEPT OF BIOCHEMISTRY UNIVERSITY OF ALLAHABAD 7. Dr. ROSHAN ALAM (1958) ALLAHABAD 211002 C/O MOHD HABIB EDUCATIONAL HOUSE AMU SHAM SHAD MARKET 16. -

Human RNA Viruses 10–12 February, 2010
ICGEB-IUBMB Workshop on Human RNA Viruses 10–12 February, 2010 Co-Organizers Shahid Jameel, ICGEB, New Delhi, India Navin Khanna, ICGEB, New Delhi, India Alessandro Marcello, ICGEB, Trieste, Italy Oscar Burrone, ICGEB, Trieste, Italy Organized by International Centre for Genetic Engineering and Biotechnology Aruna Asaf Ali Marg, New Delhi-110067, India First Impression: 2010 © ICGEB, New Delhi ISBN: 978-93-80043-77-7 No part of this publication may be reproduced or transmitted in any form by any means, electronic or mechanical, including photocopy, recording, or any information storage and retrieval system, without permission in writing from the copyright owners. DISCLAIMER The authors are solely responsible for the contents of the papers compiled in this volume. The publishers or editors do not take any responsibility for the same in any manner. Errors, if any, are purely unintentional and readers are requested to communicate such errors to the editors or publishers to avoid discrepancies in future. Published by EXCEL INDIA PUBLISHERS 61/28, Dalpat Singh Building, Pratik Market, Munirka, New Delhi-110067 Tel: +91-11-2671 1755/ 2755/ 5755 z Fax: +91-11-2671 6755 E-mail: [email protected] Website: www.excelpublish.com Typeset by Excel Publishing Services, New Delhi - 110067 E-mail: [email protected] Printed by Excel Seminar Solutions, New Delhi - 110067 E-mail: [email protected] II Workshop “Human RNA Viruses” 10th February – 12th February 2010 ICGEB New Delhi PROGRAMME Wednesday 10th February 08:00 – Departure from Hotels 08:30 – Registration 09:30 – Inaugural session (Director, ICGEB, New Delhi and Workshop coordinators) Session 1 10:00 – 10:45 Dr. -

List of Life Members As on 1St Frbruary 2018
LIST OF LIFE MEMBERS AS ON 1ST FRBRUARY 2018 ALIGARH 10. Dr. SHAGUFTA MOIN (1261) DEPT. OF BIOCHEMISTRY 1. Dr. HAMMAD AHMAD SHADAB G. G.(1454) J. N. MEDICAL COLLEGE 31 SECTOR OF GENETICS ALIGARH MUSLIM UNIVERSITY DEPT. OF ZOOLOGY ALIGARH 202 002 ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 11. SHAIK NISAR ALI (3769) DEPT. OF BIOCHEMISTRY 2. Dr. INDU SAXENA (1838) FACULTY OF LIFE SCIENCE HIG 30, ADA COLONY ALIGARH MUSLIM UNIVERSITY AVANTEKA PHASE I ALIGARH 202 002 RAMGHAT ROAD, ALIGARH 202 001 [email protected] 3. Dr. (MRS) KHUSHTAR ANWAR SALMAN(3332) ALLAHABAD DEPT. OF BIOCHEMISTRY JAWAHARLAL NEHRU MEDICAL COLLEGE 12. Dr. ABHAY KUMAR PANDEY (2054) ALIGARH MUSLIM UNIVERSITY DEPT OF BIOCHEMISTRY ALIGARH 202 002 UNIVERSITY OF ALLAHABAD [email protected] ALLAHABAD 211002 4. Dr. MOHAMMAD TABISH (2226) 13. Dr. AMIT KUMAR (2123) DEPT. OF BIOCHEMISTRY DEPT. OF ZOOLOGY FACULTY OF LIFE SCIENCES ALLAHABAD UNIVERSITY ALIGARH MUSLIM UNIVERSITY ALLAHABAD 211 002 ALIGARH 202 002 [email protected] 14. Dr. AMITA MISHRA (2665) C/O SRI RAMESH MISHRA 5. Dr. MOHAMMAD AFZAL (1101) G B 53, GANGA SECTOR, TRIVENIPURAM DEPT. OF ZOOLOGY JHUSI , ALLAHABAD 211 019 ALIGARH MUSLIM UNIVERSITY [email protected] ALIGARH 202 002 15. Dr. ANJANA PANDEY (1549) 6. Dr. RIAZ AHMAD(1754) CENTRE FOR BIOTECHNOLOGY SECTION OF GENETICS MOTILAL NEHRU NATIONAL INSTITUTE OF DEPT. OF ZOOLOGY TECHNOLOGY ALIGARH MUSLIM UNIVERSITY ALLAHABAD 211 004 ALIGARH 202 002 16. ASHUTOSH GUPTA (3789) 7. Dr. ROSHAN ALAM (1958) 116A/7B JAYANTIPUR C/O MOHD HABIB EDUCATIONAL HOUSE SULEM SARAI AMU SHAM SHAD MARKET ALLAHABAD 211 011 ALIGARH 202 002 [email protected] 8. -

SBC(I) PRESIDENTS Dr. G. J. Fowler 1931, 1946-1948 Dr. Upendranath Brahmachari 1932, 1934-1935 Shri S. L. Bhatia 1933, 1949
SBC(I) PRESIDENTS Dr. G. J. Fowler 1931, 1946-1948 Dr. Upendranath Brahmachari 1932, 1934-1935 Shri S. L. Bhatia 1933, 1949 Dr. V. Surbahmanyan 1950-1951 Dr. N. R. Dhar 1952-1953 Dr. B. Sanjiva Rao 1954-1955 Dr. B. C. Guha 1956-1957 Dr. K. V. Giri 1958 Dr. D. K. Banerjee 1958 Dr. V. R. Khanolkar 1959-1961 Dr. P. S. Sarma 1962 -1963 Dr. A. Sreenivasan 1964-1966 Dr. B. Jagannathan 1967- 1968 Dr. B. K. Bachhawat 1969- 70,1979,1990-1994 Dr. H. R. Cama 1971-1972 Dr. A. N. Radhakrishnan 1973-1974 Dr. T.A. Venkitasubramanian 1975-1976 Dr. M. R. Raghavendra Rao 1977-1978 Dr. C. R. Krishna Murti 1979-1980 Dr. P. M. Bhargava 1981-1982 Dr. K.K.G .Menon 1983-1984 Dr. John Banabas 1985-1986 Dr. N. Appaji Rao 1987-1988 Dr. A. Salahuddin 1989-1990 Dr. G. Padmanban 1995-1996 Dr. Asis Datta 1997-1998 Dr. Amar Nath Bhaduri 1999-2000 Dr. M. R. S. Rao 2001-2004 Dr. C. M. Gupta 2005- 2006 Dr. D Balasubramanian 2007-2008 Dr. V. S. Chauhan 2009-2010 Dr. V. Nagaraja 2011-2012 Dr. Dhrubajyothi Chattopadhyay 2013- 2014 Dr. Ch. Mohan Rao 2015-2016 Dr. Umesh Varshney 2017-Current SBC(I) VICE PRESIDENTS 1991- 1992 Dr. S Mahadevan,Bangalore Dr. A N Bhaduri,Kolkata Dr K L Bajaj , Ludhiana 1993-1994 Dr K P Gopinathan,Bangalore Dr R K Mandal, Kolkata Dr. R K Jethi, Chandigarh 1995-1996 Dr. M S Shaila, Bangalore Dr. Jyotimoy Das,Kolkata Dr. G K Garg, Pantnagar 1997- 1998 Dr.