Authentication of Holy Basil Using Markers Relating to a Toxicology‑Relevant Compound
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Antimicrobial Activity of Tulsi (Ocimum Tenuiflorum) Essential Oil
fmicb-07-00681 May 12, 2016 Time: 16:34 # 1 View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Frontiers - Publisher Connector ORIGINAL RESEARCH published: 17 May 2016 doi: 10.3389/fmicb.2016.00681 Antimicrobial Activity of Tulsi (Ocimum tenuiflorum) Essential Oil and Their Major Constituents against Three Species of Bacteria Hanaa A. Yamani1,2, Edwin C. Pang1, Nitin Mantri1* and Margaret A. Deighton1 1 School of Science, Royal Melbourne Institute of Technology University, Melbourne, VIC, Australia, 2 Biology, Section Microbiology, School of Applied Sciences, King Abdulaziz University, Jeddah, Saudi Arabia In recent years scientists worldwide have realized that the effective life span of any antimicrobial agent is limited, due to increasing development of resistance by microorganisms. Consequently, numerous studies have been conducted to find new alternative sources of antimicrobial agents, especially from plants. The aims of this project were to examine the antimicrobial properties of essential oils distilled from Australian-grown Ocimum tenuiflorum (Tulsi), to quantify the volatile components present in flower spikes, leaves and the essential oil, and to investigate the compounds responsible for any activity. Broth micro-dilution was used to determine the minimum Edited by: Yuji Morita, inhibitory concentration (MIC) of Tulsi essential oil against selected microbial pathogens. Aichi Gakuin University, Japan The oils, at concentrations of 4.5 and 2.25% completely inhibited the growth Reviewed by: of Staphylococcus aureus (including MRSA) and Escherichia coli, while the same Osmar Nascimento Silva, Dom Bosco Catholic University, Brazil concentrations only partly inhibited the growth of Pseudomonas aeruginosa. Of 54 J. -

To Assess the Role of Natural Pesticides Made from Tulsi
International Journal of Applied Pharmaceutics ISSN- 0975-7058 Vol 8, Issue 2, 2016 Original Article TO ASSESS THE ROLE OF NATURAL PESTICIDES MADE FROM TULSI OCIMUM TENUIFLORUM, TURMERIC CURCUMA LONGA AND NEEM AZADIRACHTA INDICA ON CULINARY CROPS AND ITS MATURING SOIL DEEKSHA GUPTA*, SHILPA SIVADAS, KEERTHI VIKRAM, SUNEETHA V School of Bio Sciences and Technology, VIT University, Vellore 632014 Email: [email protected] Received: 10 Dec 2015, Revised and Accepted: 09 Apr 2016 ABSTRACT Objective: Our research plans on creating natural pesticides to serve the same along with keeping it sustainable, effective and healthy. Our project aims to find the effects of natural pesticides like neem, tulsi and turmeric on plants like mung bean, chickpea, methi (fenugreek) and other varieties. We also aim to provide a solution from all the other techniques that have been exhaustively implied and are being created to ensure the fulfillment for the future generations, e. g. GMO Methods: The extracts of neem, tulsi and turmeric are made by grinding and mixing it with water in 1:1 ratio and are then categorically sprayed over a range of 12 pots separated according to different types of plants namely mung, methi and channa. The growth of the plants and the soil analysis (by serial dilution) is done over the microbial growth, resistance to fungal infections and other pest infestations. Results: We came up with a positive result showing less chance of infections and decrease in the colonies of harmful bacteria and pathogens with the help of serially diluting the soil sample under the effect of natural pesticides and pore plating it on the media. -

Hort-Science-Holy-Basil-Article.Pdf
HORTSCIENCE 53(9):1275–1282. 2018. https://doi.org/10.21273/HORTSCI13156-18 To increase cultivation of holy basil in the southeastern United States, the first step is to evaluate available holy basil varieties to de- Variation in Growth and Development, termine which are most suited for commer- cial production. At present, growers typically and Essential Oil Yield between Two select varieties based on seed availability, market demand, and harvestable weight, and Ocimum Species (O. tenuiflorum and not necessarily on the presence or concentra- tion of biologically active compounds (Zhang et al., 2012). With medicinal herbs, O. gratissimum) Grown in Georgia an important consideration is the measurable Noelle J. Fuller1 difference in therapeutic constituents, such as Department of Horticulture, University of Georgia, 1111 Miller Plant essential oils, that are indicators of quality and efficacy. For example, a notable phenolic Sciences Building, Athens, GA 30602 compound found in holy basil essential oil is Ronald B. Pegg eugenol. It is a versatile molecule with application in many industries (Kamatou Department of Food Science and Technology, University of Georgia, 100 et al., 2012). It has a spicy clove-like scent Cedar Street, Athens, GA 30602 and has been shown to be therapeutically effective for neurological, inflammatory, al- James Affolter lergic, and immunological disorders (Bakkali State Botanical Garden of Georgia, 450 South Milledge, Athens, GA 30605 et al., 2008; Kamatou et al., 2012; Sen, 1993). Eugenol is largely extracted from natural David Berle sources, most commonly clove essential oil Department of Horticulture, University of Georgia, 1111 Miller Plant (Eugenia caryophyllata), which has a gross Sciences Building, Athens, GA 30602 market value of US$30–70 million annually for use in food and cosmetics (Bohnert et al., Additional index words. -

1. Basil 'Cardinal' 2
HERBS 1. BASIL 'CARDINAL' 2. BASIL 'DOLCE FRESCA' 3. BASIL ‘GENOVESE’ 4. BASIL ‘LEMON’ 5. BASIL ‘PROSPERA ITALIAN LARGE LEAF’ 6. BASIL 'SWEET THAI' 7. BASIL ‘THAI HOLY KAPRAO’ 8. CHIVES 'ONION' 9. CILANTRO 'SANTO' 10. DILL ‘BOUQUET’ 11. FENNEL ‘BRONZE’ 12. FENNEL ‘GROSFRUCHTIGER’ 13. MARJORAM 'SWEET' 14. OREGANO 'ITALIAN' 15. PARSLEY ‘GIANT OF ITALY’ 16. PARSLEY 'TRIPLE CURLED' 17. ROSEMARY ARP 18. SAGE ‘PURPLE’ 19. THYME ‘LEMON’ Basil ‘Cardinal’ Height: 24 - 30" Exposure: Sun General Information: At first glance you’ll think this is a celosia, with its heavy, tightly packed blooms. Strong, deep burgundy stems hold the flowers above the smooth, bright green leaves, ensuring a vibrant show throughout the summer even as you continue harvesting fresh leaves. Basil ‘Dolce Fresca’ Plant Habit: Mounded Spacing: 10 - 12" Height: 12 - 14" Exposure: Sun General Information: Tidy plant with an abundance of savory leaves that can be used in any dish calling for basil. Grows very well in a container or in the ground. Mid-size plant has a unique bushy habit with shorter internodes that holds its controlled size in the garden without getting tall and leggy. Basil ‘Genovese’ Classic Italian variety. Authentic flavor and appearance. Tall and relatively slow to bolt with large dark-green leaves about 3" long. Ht. 24-30". •Edible Flowers: Use the flowers in any recipe that calls for basil, or to garnish drinks, salads, soups, pasta, and desserts. Flavor is of intense basil. Basil ‘Lemon’ Lemony aroma and flavor. Attractive, spreading silver-green plant with lemony aroma and flavor is great for potpourris, tea, chicken, fish, vegetables and herb vinegars. -

Morphological Characteristics and Susceptibility of Basil Species and Cultivars to Peronospora Belbahrii
DISEASE AND PEST MANAGEMENT HORTSCIENCE 51(11):1389–1396. 2016. doi: 10.21273/HORTSCI09778-16 P. Belbahrii, was first reported in Uganda in 1932 as Peronospora sp. and later in 1937 as Peronospora lamii (Hansford, 1933, Morphological Characteristics and 1938). BDM was not reported again until 2001 in Switzerland (Heller and Baroffio, Susceptibility of Basil Species and 2003). After this initial confirmation, other countries throughout Europe, the Mediter- Cultivars to Peronospora belbahrii ranean, and continents across the world reported BDM for the first time (Belbahri Kathryn Homa et al., 2005; Kofoet et al., 2008; Lefort et al., Department of Plant Biology and Pathology, Rutgers University, Foran Hall, 59 2008; McGrath, 2011). This pathogen was Dudley Road, New Brunswick, NJ 08901-8520; and IR-4 Project Headquarters, first reported in the United States in 2007 in southern Florida (Roberts et al., 2009). Rutgers University, 500 College Road East, Suite 201W, Princeton 08540 Since then, the disease has spread across the William P. Barney continental United States and Hawaii (Wye- nandt et al., 2015). Although the epidemiol- IR-4 Project Headquarters, Rutgers University, 500 College Road East, Suite ogy of the pathogen is not completely 201W, Princeton 08540 understood, BDM appears to have been spread globally via infested seed as well as Daniel L. Ward and Christian A. Wyenandt through wind currents (Thines et al., 2009; Department of Plant Biology and Pathology, Rutgers University, Rutgers Wyenandt et al., 2015). Agricultural Research and Extension Center, 121 Northville Road, Over 5000 ha of sweet basil is grown in Bridgeton, NJ 08302 the United States on an annual basis (J.E. -

Phcogj.Com Ocimum Sanctum: Role in Diseases Management Through
Pharmacogn J. 2020; 12(5): 1198-1205 A Multifaceted Journal in the field of Natural Products and Pharmacognosy Review Article www.phcogj.com Ocimum sanctum: Role in Diseases Management Through Modulating Various Biological Activity Saleh A. Almatroodi1, Mohammed A. Alsahli1, Ahmad Almatroudi1, Arshad Husain Rahmani1,* ABSTRACT Medicinal plants are used commonly by traditional medical practitioners in their daily practice for the treatment of various diseases. The treatment based on natural products are preferred because they are more economic and have lesser side-effects. In this regards, Ocimum sanctum 1 Saleh A. Almatroodi , commonly known as holy basil or tulsi is used in the diseases cure and treatment since ancient 1 Mohammed A. Alsahli , Ahmad time. Ocimum sanctum has been proven health promoting effect through modulation of Almatroudi1, Arshad Husain various biological activates. Ocimum sanctum shows therapeutic role through its anti-oxidant, 1, Rahmani * anti-inflammatory, anti-microbial, anti-diabetic, hepatoprotective and wound healing effects. Department of Medical Laboratories, College Besides, the constituents of holy basil have been confirmed to have a noteworthy effect in of Applied Medical Sciences, Buraydah cancer management through inhibition of cancer development and progression. Further, the 52571, Qassim University, SAUDI ARABIA. synergistic effect of Ocimum sanctum component with anti-cancer drugs has been proven as it reduces the growth of cancer. Molecular mechanism and human clinical trials based should Correspondence be performed to avail its role in diseases cure and management. This review comprehensively Arshad Husain Rahmani summarizes the role of holy basil in disease management through in vivo and in vitro study. Department of Medical Laboratories, College of Applied Medical Sciences, Key words: Ocimum sanctum, Health promoting effect, Anti-cancer, Anti-oxidant effect. -

Holy Basil Ocimum Tenuiflorum
Did You Know? Holy Basil Ocimum tenuiflorum • Additional common names include tulsi, tulasi, and sacred basil. • In its native India, holy basil is particularly sacred herb in the Hindu tradition where it is thought to be the manifestation of the goddess, Tulasi, and to have grown from her ashes. • In one version of the legend, Tulasi was tricked into betraying her husband when she was seduced by the god Vishnu in the guise of her husband. In her torment, Tulasi killed herself, and Vishnu declared that she would be “worshipped by women for her faithfulness” and would keep women from becoming widows. • Holy basil, also referred to as tulsi basil in reference to the goddess Tulasi, became the symbol of love, eternal life, purification and protection. • Holy basil has also played a role in burial rituals, including scattering the leaves on graves as well as growing the plant on graves. • There are a few species and varieties referred to as holy basil and all are in the same genus as common garden basil. • Like other basils, holy basil is a member of the mint family (Lamiaceae). • Historical medicinal uses include treatment of colds and flu due to its antiviral, antibacterial, decongestant and diaphoretic properties. In India, it is used in a tea to clear congestion. • Other medicinal uses are said to include immune strengthening and balancing, balancing blood sugar, stimulating appetite, soothing digestion and relieving insect stings. ©2016 by The Herb Society of America www.herbsociety.org 440-256-0514 9019 Kirtland Chardon Road, Kirtland, OH 44094. -
A Study on Antidermatophytic Potential of Ocimum Tenuiflorum Essential Oil and Chemical Composition Evaluation
International Journal of PharmTech Research CODEN (USA): IJPRIF, ISSN: 0974-4304, ISSN(Online): 2455-9563 Vol.9, No.11, pp 151-160, 2016 A study on Antidermatophytic Potential of Ocimum tenuiflorum Essential Oil and Chemical Composition Evaluation Vishnu Sharma, Anima Sharma* and Ruchi Seth Department of Biotechnology, JECRC University, Jaipur, India Abstract : The Dermatophytes engage in an important role in the atmosphere. They invade the keratinophilic substrates with causing the superficial infections. The certain augmentation in the infection in the human, there is urgent need to search out a new therapeutics or remedies from nature. In the presented study, the chemical composition of Ocimum tenuiflorum’s volatile oil and its anti-dermatophytic potential was evaluated against isolated Dermatophyte species from Jaipur (India). The Clear pale yellowish colored oil containing 40 volatile components was extracted & analyzed using Hydro Distillation Process and GC & GC-MS methods. The main constitutions were obtained as β-Caryophyllene (38.90%) and Eugenol (19.63%)in the oil and it revealed excellent inhibition activity against test fungal organisms with presence of Maximum Inhibition Zone of 37 mm against T. mentagrophytes(KU578106)as well as 31.67 mm for M. gypsiumand 28.33mmfor M. nannum as compared to standard. The results studied in the present research, helps to look out new natural therapeutic drugs from Ocimum tenuiflorum as an anti-dermatophytic agent than the standard. Keywords: Dermatophytes; Ocimum tenuiflorum; Volatile; Hydro Distillation; β-Caryophyllene. Introduction: Fungi have renowned as a unique division in nature from evolution1. In fungi divisions, the Deuteromycota Division has awide account of pathogenic fungi in which some have the ability to penetrate the natural keratin and play a role as keratinolytic agent2-3. -

Journal of Pharmacognocy & Phytochemistry
e-ISSN: 2321-6182 p-ISSN: 2347-2332 Research and Reviews: Journal of Pharmacognocy & Phytochemistry Medicinal plant Tulasi and its Uses P. Udaya Lakshmi* Montessori shiva shivani college of Pharmacy, Guntur. Short Commentary Article Received: 04/05/2015 Revised: 27/05/2015 *For Correspondence Accepted: 02/06/2015 P. Udaya Lakshmi, Montessori shiva shivani college of Pharmacy, Guntur, Tel: 8125171946; E-mail: [email protected] . Keywords: Ocimum tenuiflorum, tinea, Tulsi Introduction Ocimum tenuiflorum, also called Ocimum sanctum, holy basil, or tulasi, is associate degree aromatic plant within the Labiatae that is native to the Indian landmass and widespread as a tracheophyte throughout the Southeast Asian tropics.[1-5] it's an erect, several branched suffrage, 30–60 cm tall with furry stems and straightforward opposite inexperienced or purple leaves that area unit powerfully scented. Leaves have petioles and area unit ovate, up to five cm long, typically slightly toothed. The flowers area unit violet in elongate racemes in shut whorls. The 2 main morphotypes cultivated in India and Kingdom of Nepal area unit green-leaved (Sri or Hindu deity tulasi) and purple-leaved (Krishna tulasi) The holy basil is additionally a flavoring remedy for lots of common ailments. Here're prime fifteen healthful uses of tulsi. 1. Healing Power: The tulsi plant has many medicinal properties. The leaves area unit a nerve tonic and additionally sharpen memory. [6] 2. Fever & Common Cold: The leaves of basil square measure specific for several fevers. Throughout the season, once protozoal infection and dengue square measure wide prevailing, tender leaves, cooked with tea, act as preventive against these diseases. -

Herb & Flower Plant List
2317 Evergreen Rd. Herb & Flower Louisa, Va 23093 (540)967-1165 (434)882-2648 Plant List www.forrestgreenfarm.com COMMON NAME : LATIN NAME COMMON NAME : LATIN NAME COMMON NAME : LATIN NAME Ageratum, Blue Planet : Burdock, Chiko : Arctium lappa Echinacea, Primadonna Deep Rose : Echinacea purpurea Agrimony : Agrimonia eupatoria Calendula : Calendula officinalis Echinacea, Primadonna White : Echinacea purpurea Amaranth, burgundy : Amaranth sp. Calendula, HoriSun Yellow : Calendula officinalis Echinacea, Purpurea : Amaranthus Love-lies-Bleeding : An=maranthus caudatusCalendula, Solis Sponsa : Calendula Officinalis Elderberry : Sambucus nigra Andrographis : Andrographis paniculata Calibrachoa, Royal Purple : Elderberry, Adams : Sambucus canadensis spp. Angelica : Angelica archangelica Calibrachoa, Ultimate Pink : Elderberry, Eridu : Sambucus canadensis spp. Angelonia, blue : Castor Bean : Ricinus communis Elderberry, Magnolia : Sambucus canadensis spp. Anise : pimpinella anisum Catmint : Nepeta mussinii Elderberry, Ranch : Sambucus canadensis spp. Anise-Hyssop : Agastache foeniculum Catnip : Nepeta Cataria Elderberry, Wyldewood : Argyranthemum : Butterfly Yellow Celandine : Chelidonium majus Elecampane : Inula helenium Artichoke, Imperial Star : Cynara scolymus Celosia, Cramer's Burgundy : Elephant Ear : Colocasia gigantea Asclepias : Asclepias curassavica Celosia, Cramer's Rose : Evening Primrose : Oenothera biennis Ashitaba : Angelica keiskei koidzumi Chamomile, Common (German) : Matricaria recutita Everlasting : Helichrysum Ashwagandha -

Ocimum Tenuiflorum – Holy Basil
New! Ocimum tenuiflorum – Holy Basil Research Summary Ocimum tenuiflorum 1:2 Fluid extract Holy Basil is native to tropical Asia and though relatively Synonyms: Ocimum sanctum new to western medicine systems, has been grown and used medicinally in India for over 3000 years. Also Common Names: Holy Basil //Tulsi // Surasa // Tulasi // known as ‘Tulsi’, it is one of the most sacred plants in Karuttulaci // Tulaci India & it has the status of deity on the Indian subcontinent. Botanical family: Lamiaceae There is a growing body of research into a wide range Part Used: Dried Leaf of possible therapeutic benefits of Holy Basil, much of which suggests protective actions against health concerns common in today’s environment. These Dosage: 20-50ml per week properties make it an invaluable component of the Eugenol // Ursolic acid // modern herbalists toolkit. Primary Active Constituents: Rosmarininc acid // α & β-caryophyllene // β-elemene // Anti-aging germacrene D // Ocimumoside A & B // Flavonoids (orientin & vicenin) // Oleanolic acid Anti-inflammatory & antioxidant actions have been demonstrated in a number of studies, & the Contraindications: Pregnancy & lactation. components eugenol & ursolic acid are believed to May act as a male contraceptive agent in large doses. significantly contribute to these actions. Ursolic acid is also well established as an anti-aging agent, helping to Actions: Adaptogen // Neuroprotective // Antioxidant; // maintain healthy skin and promoting skin elasticity. Chemoprotective // Nervine // Immune modulator // Anti- Ocimum extract also extended the lifespan of the fruit fly Caenorhabditis elegans, used as a genetic model for inflammatory // Hepatoprotective // Anti-microbial studies of aging and the role of stress (Ishii 2005; Main Indications: Pandey 2013). -
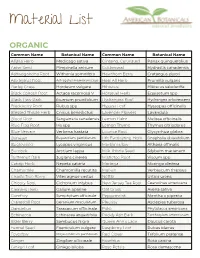
Material List
Material List ORGANIC Common Name Botanical Name Common Name Botanical Name Alfalfa Herb Medicago sativa Ginseng, Cultivated Panax quinquefolius Anise Seed Pimpinella anisum Goldenseal Hydrastis canadensis Ashwagandha Root Withania somnifera Hawthorn Berry Crataegus piperi Astragalus Root Astragalus membranaceus Heal All Herb Prunella vulgaris Barley Grass Hordeum vulgare Hibiscus Hibiscus sabdariffa Black Cohosh Root Actaea racemosa V Horsetail Herb Equisetum spp Black Haw Bark iburnum prunifolium Hydrangea Root Hydrangea arborescens Blackberry Root Rubus spp Hyssop Leaf Hyssopus officinalis Blessed Thistle Herb Cnicus benedictus Lavender Flowers Lavandula Blood Root Sanguinaria canadensis Lemon Balm Melissa officinalis Blue Flag Root Iris spp Lemon Thyme Thymus citriodorus Blue Vervain Verbena hastata Licorice Root Glycyrrhiza glabra Boneset Eupatorium perfoliatum Life Everlasting Herb Gnaphaliu obiusifolium Bugleweed Lycopus virginicus Marshmallow Althaea offinalis Burdock Arctium lappa Milk Thistle Seed Silybum marianum Butternut Bark Juglans cinerea Mistletoe Root Viscum spp Catnip Herb Nepeta cataria Moringa Moringa oleifera Chamomile Chamomilla recutita Mullein Verbascum thapsus Chaste Tree Berry Vitex agnus-castus Nettle Urtica urens Chicory Root Cichorium intybus New Jersey Tea Root Ceanothus americana Cleavers Herb Galium aparine Oat Grass Avena sativa Comfrey Symphytum officinale Peppermint Mentha x piperita Cranesbill Root Geranium maculatum Pleurisy Asclepias tuberosa Dandelion Taraxacum officinale Poke Phytolacca americana