Bayesian Nonparametric Regression for Educational Research
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

On Assessing Binary Regression Models Based on Ungrouped Data
Biometrics 000, 1{18 DOI: 000 000 0000 On Assessing Binary Regression Models Based on Ungrouped Data Chunling Lu Division of Global Health, Brigham and Women's Hospital & Department of Global Health and Social Medicine Harvard University, Boston, U.S. email: chunling [email protected] and Yuhong Yang School of Statistics, University of Minnesota, Minnesota, U.S. email: [email protected] Summary: Assessing a binary regression model based on ungrouped data is a commonly encountered but very challenging problem. Although tests, such as Hosmer-Lemeshow test and le Cessie-van Houwelingen test, have been devised and widely used in applications, they often have low power in detecting lack of fit and not much theoretical justification has been made on when they can work well. In this paper, we propose a new approach based on a cross validation voting system to address the problem. In addition to a theoretical guarantee that the probabilities of type I and II errors both converge to zero as the sample size increases for the new method under proper conditions, our simulation results demonstrate that it performs very well. Key words: Goodness of fit; Hosmer-Lemeshow test; Model assessment; Model selection diagnostics. This paper has been submitted for consideration for publication in Biometrics Goodness of Fit for Ungrouped Data 1 1. Introduction 1.1 Motivation Parametric binary regression is one of the most widely used statistical tools in real appli- cations. A central component in parametric regression is assessment of a candidate model before accepting it as a satisfactory description of the data. In that regard, goodness of fit tests are needed to reveal significant lack-of-fit of the model to assess (MTA), if any. -

Nonparametric Regression 1 Introduction
Nonparametric Regression 10/36-702 Larry Wasserman 1 Introduction Now we focus on the following problem: Given a sample (X1;Y1);:::,(Xn;Yn), where d Xi 2 R and Yi 2 R, estimate the regression function m(x) = E(Y jX = x) (1) without making parametric assumptions (such as linearity) about the regression function m(x). Estimating m is called nonparametric regression or smoothing. We can write Y = m(X) + where E() = 0. This follows since, = Y − m(X) and E() = E(E(jX)) = E(m(X) − m(X)) = 0 A related problem is nonparametric prediction. Given a pair (X; Y ), we want to predict Y from X. The optimal predictor (under squared error loss) is the regression function m(X). Hence, estimating m is of interest for its own sake and for the purposes of prediction. Example 1 Figure 1 shows data on bone mineral density. The plots show the relative change in bone density over two consecutive visits, for men and women. The smooth estimates of the regression functions suggest that a growth spurt occurs two years earlier for females. In this example, Y is change in bone mineral density and X is age. Example 2 Figure 2 shows an analysis of some diabetes data from Efron, Hastie, Johnstone and Tibshirani (2004). The outcome Y is a measure of disease progression after one year. We consider four covariates (ignoring for now, six other variables): age, bmi (body mass index), and two variables representing blood serum measurements. A nonparametric regression model in this case takes the form Y = m(x1; x2; x3; x4) + . -

Nonparametric Regression with Complex Survey Data
CHAPTER 11 NONPARAMETRIC REGRESSION WITH COMPLEX SURVEY DATA R. L. Chambers Department of Social Statistics University of Southampton A.H. Dorfman Office of Survey Methods Research Bureau of Labor Statistics M.Yu. Sverchkov Department of Statistics Hebrew University of Jerusalem June 2002 11.1 Introduction The problem considered here is one familiar to analysts carrying out exploratory data analysis (EDA) of data obtained via a complex sample survey design. How does one adjust for the effects, if any, induced by the method of sampling used in the survey when applying EDA methods to these data? In particular, are adjustments to standard EDA methods necessary when the analyst's objective is identification of "interesting" population (rather than sample) structures? A variety of methods for adjusting for complex sample design when carrying out parametric inference have been suggested. See, for example, SHS, Pfeffermann (1993) and Breckling et al (1994). However, comparatively little work has been done to date on extending these ideas to EDA, where a parametric formulation of the problem is typically inappropriate. We focus on a popular EDA technique, nonparametric regression or scatterplot smoothing. The literature contains a limited number of applications of this type of analysis to survey data, usually based on some form of sample weighting. The design-based theory set out in chapter 10, with associated references, provides an introduction to this work. See also Chesher (1997). The approach taken here is somewhat different. In particular, it is model-based, building on the sample distribution concept discussed in section 2.3. Here we develop this idea further, using it to motivate a number of methods for adjusting for the effect of a complex sample design when estimating a population regression function. -

Nonparametric Bayesian Inference
Nonparametric Bayesian Methods 1 What is Nonparametric Bayes? In parametric Bayesian inference we have a model M = ff(yjθ): θ 2 Θg and data Y1;:::;Yn ∼ f(yjθ). We put a prior distribution π(θ) on the parameter θ and compute the posterior distribution using Bayes' rule: L (θ)π(θ) π(θjY ) = n (1) m(Y ) Q where Y = (Y1;:::;Yn), Ln(θ) = i f(Yijθ) is the likelihood function and n Z Z Y m(y) = m(y1; : : : ; yn) = f(y1; : : : ; ynjθ)π(θ)dθ = f(yijθ)π(θ)dθ i=1 is the marginal distribution for the data induced by the prior and the model. We call m the induced marginal. The model may be summarized as: θ ∼ π Y1;:::;Ynjθ ∼ f(yjθ): We use the posterior to compute a point estimator such as the posterior mean of θ. We can also summarize the posterior by drawing a large sample θ1; : : : ; θN from the posterior π(θjY ) and the plotting the samples. In nonparametric Bayesian inference, we replace the finite dimensional model ff(yjθ): θ 2 Θg with an infinite dimensional model such as Z F = f : (f 00(y))2dy < 1 (2) Typically, neither the prior nor the posterior have a density function with respect to a dominating measure. But the posterior is still well defined. On the other hand, if there is a dominating measure for a set of densities F then the posterior can be found by Bayes theorem: R A Ln(f)dπ(f) πn(A) ≡ P(f 2 AjY ) = R (3) F Ln(f)dπ(f) Q where A ⊂ F, Ln(f) = i f(Yi) is the likelihood function and π is a prior on F. -
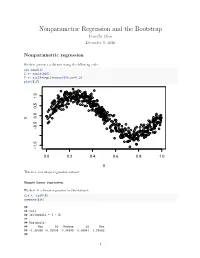
Nonparametric Regression and the Bootstrap Yen-Chi Chen December 5, 2016
Nonparametric Regression and the Bootstrap Yen-Chi Chen December 5, 2016 Nonparametric regression We first generate a dataset using the following code: set.seed(1) X <- runif(500) Y <- sin(X*2*pi)+rnorm(500,sd=0.2) plot(X,Y) 1.0 0.5 0.0 Y −0.5 −1.5 0.0 0.2 0.4 0.6 0.8 1.0 X This is a sine shape regression dataset. Simple linear regression We first fit a linear regression to this dataset: fit <- lm(Y~X) summary(fit) ## ## Call: ## lm(formula = Y ~ X) ## ## Residuals: ## Min 1Q Median 3Q Max ## -1.38395 -0.35908 0.04589 0.38541 1.05982 ## 1 ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 1.03681 0.04306 24.08 <2e-16 *** ## X -1.98962 0.07545 -26.37 <2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 '' 1 ## ## Residual standard error: 0.4775 on 498 degrees of freedom ## Multiple R-squared: 0.5827, Adjusted R-squared: 0.5819 ## F-statistic: 695.4 on 1 and 498 DF, p-value: < 2.2e-16 We observe a negative slope and here is the scatter plot with the regression line: plot(X,Y, pch=20, cex=0.7, col="black") abline(fit, lwd=3, col="red") 1.0 0.5 0.0 Y −0.5 −1.5 0.0 0.2 0.4 0.6 0.8 1.0 X After fitted a regression, we need to examine the residual plot to see if there is any patetrn left in the fit: plot(X, fit$residuals) 2 1.0 0.5 0.0 fit$residuals −0.5 −1.0 0.0 0.2 0.4 0.6 0.8 1.0 X We see a clear pattern in the residual plot–this suggests that there is a nonlinear relationship between X and Y. -

Nonparametric Regression
Nonparametric Regression Statistical Machine Learning, Spring 2015 Ryan Tibshirani (with Larry Wasserman) 1 Introduction, and k-nearest-neighbors 1.1 Basic setup, random inputs Given a random pair (X; Y ) Rd R, recall that the function • 2 × f0(x) = E(Y X = x) j is called the regression function (of Y on X). The basic goal in nonparametric regression is ^ d to construct an estimate f of f0, from i.i.d. samples (x1; y1);::: (xn; yn) R R that have the same joint distribution as (X; Y ). We often call X the input, predictor,2 feature,× etc., and Y the output, outcome, response, etc. Importantly, in nonparametric regression we do not assume a certain parametric form for f0 Note for i.i.d. samples (x ; y );::: (x ; y ), we can always write • 1 1 n n yi = f0(xi) + i; i = 1; : : : n; where 1; : : : n are i.i.d. random errors, with mean zero. Therefore we can think about the sampling distribution as follows: (x1; 1);::: (xn; n) are i.i.d. draws from some common joint distribution, where E(i) = 0, and then y1; : : : yn are generated from the above model In addition, we will assume that each i is independent of xi. As discussed before, this is • actually quite a strong assumption, and you should think about it skeptically. We make this assumption for the sake of simplicity, and it should be noted that a good portion of theory that we cover (or at least, similar theory) also holds without the assumption of independence between the errors and the inputs 1.2 Basic setup, fixed inputs Another common setup in nonparametric regression is to directly assume a model • yi = f0(xi) + i; i = 1; : : : n; where now x1; : : : xn are fixed inputs, and 1; : : : are still i.i.d. -

A Weakly Informative Default Prior Distribution for Logistic and Other
The Annals of Applied Statistics 2008, Vol. 2, No. 4, 1360–1383 DOI: 10.1214/08-AOAS191 c Institute of Mathematical Statistics, 2008 A WEAKLY INFORMATIVE DEFAULT PRIOR DISTRIBUTION FOR LOGISTIC AND OTHER REGRESSION MODELS By Andrew Gelman, Aleks Jakulin, Maria Grazia Pittau and Yu-Sung Su Columbia University, Columbia University, University of Rome, and City University of New York We propose a new prior distribution for classical (nonhierarchi- cal) logistic regression models, constructed by first scaling all nonbi- nary variables to have mean 0 and standard deviation 0.5, and then placing independent Student-t prior distributions on the coefficients. As a default choice, we recommend the Cauchy distribution with cen- ter 0 and scale 2.5, which in the simplest setting is a longer-tailed version of the distribution attained by assuming one-half additional success and one-half additional failure in a logistic regression. Cross- validation on a corpus of datasets shows the Cauchy class of prior dis- tributions to outperform existing implementations of Gaussian and Laplace priors. We recommend this prior distribution as a default choice for rou- tine applied use. It has the advantage of always giving answers, even when there is complete separation in logistic regression (a common problem, even when the sample size is large and the number of pre- dictors is small), and also automatically applying more shrinkage to higher-order interactions. This can be useful in routine data analy- sis as well as in automated procedures such as chained equations for missing-data imputation. We implement a procedure to fit generalized linear models in R with the Student-t prior distribution by incorporating an approxi- mate EM algorithm into the usual iteratively weighted least squares. -

Testing for No Effect in Nonparametric Regression Via Spline Smoothing Techniques
Ann. Inst. Statist. Math. Vol. 46, No. 2, 251-265 (1994) TESTING FOR NO EFFECT IN NONPARAMETRIC REGRESSION VIA SPLINE SMOOTHING TECHNIQUES JUEI-CHAO CHEN Graduate Institute of Statistics and Actuarial Science, Feng Chic University, Taiehung ~072~, Taiwan, ROC (Received June 8, 1992; revised August 2, 1993) Abstract. We propose three statistics for testing that a predictor variable has no effect on the response variable in regression analysis. The test statistics are integrals of squared derivatives of various orders of a periodic smoothing spline fit to the data. The large sample properties of the test statistics are investigated under the null hypothesis and sequences of local alternatives and a Monte Carlo study is conducted to assess finite sample power properties. Key words and phrases: Asymptotic distribution, local alternatives, nonpara- metric regression, Monte Carlo. 1. Introduction Regression analysis is used to study relationships between the response vari- able and a predictor variable; therefore, all of the inference is based on the as- sumption that the response variable actually depends on the predictor variable. Testing for no effect is the same as checking this assumption. In this paper, we propose three statistics for testing that a predictor variable has no effect on the response variable and derive their asymptotic distributions. Suppose we have an experiment which yields observations (tin,y1),..., (tn~, Yn), where yj represents the value of a response variable y at equally spaced values tjn = (j - 1)/n, j = 1,...,n, of the predictor variable t. The regression model is (1.1) yj = #(tin) + ey, j --- 1,...,n, where the ej are iid random errors with E[el] = 0, Var[r ~2 and 0 K E[e 41 K oe. -

Measures of Fit for Logistic Regression Paul D
Paper 1485-2014 SAS Global Forum Measures of Fit for Logistic Regression Paul D. Allison, Statistical Horizons LLC and the University of Pennsylvania ABSTRACT One of the most common questions about logistic regression is “How do I know if my model fits the data?” There are many approaches to answering this question, but they generally fall into two categories: measures of predictive power (like R-square) and goodness of fit tests (like the Pearson chi-square). This presentation looks first at R-square measures, arguing that the optional R-squares reported by PROC LOGISTIC might not be optimal. Measures proposed by McFadden and Tjur appear to be more attractive. As for goodness of fit, the popular Hosmer and Lemeshow test is shown to have some serious problems. Several alternatives are considered. INTRODUCTION One of the most frequent questions I get about logistic regression is “How can I tell if my model fits the data?” Often the questioner is expressing a genuine interest in knowing whether a model is a good model or a not-so-good model. But a more common motivation is to convince someone else--a boss, an editor, or a regulator--that the model is OK. There are two very different approaches to answering this question. One is to get a statistic that measures how well you can predict the dependent variable based on the independent variables. I’ll refer to these kinds of statistics as measures of predictive power. Typically, they vary between 0 and 1, with 0 meaning no predictive power whatsoever and 1 meaning perfect predictions. -

Chapter 5 Nonparametric Regression
Chapter 5 Nonparametric Regression 1 Contents 5 Nonparametric Regression 1 1 Introduction . 3 1.1 Example: Prestige data . 3 1.2 Smoothers . 4 2 Local polynomial regression: Lowess . 4 2.1 Window-span . 6 2.2 Inference . 7 3 Kernel smoothing . 9 4 Splines . 10 4.1 Number and position of knots . 11 4.2 Smoothing splines . 12 5 Penalized splines . 15 2 1 Introduction A linear model is desirable because it is simple to fit, it is easy to understand, and there are many techniques for testing the assumptions. However, in many cases, data are not linearly related, therefore, we should not use linear regression. The traditional nonlinear regression model fits the model y = f(Xβ) + 0 where β = (β1, . βp) is a vector of parameters to be estimated and X is the matrix of predictor variables. The function f(.), relating the average value of the response y on the predictors, is specified in advance, as it is in a linear regression model. But, in some situa- tions, the structure of the data is so complicated, that it is very difficult to find a functions that estimates the relationship correctly. A solution is: Nonparametric regression. The general nonparametric regression model is written in similar manner, but f is left unspecified: y = f(X) + = f(x1,... xp) + Most nonparametric regression methods assume that f(.) is a smooth, continuous function, 2 and that i ∼ NID(0, σ ). An important case of the general nonparametric model is the nonparametric simple regres- sion, where we only have one predictor y = f(x) + Nonparametric simple regression is often called scatterplot smoothing, because an important application is to tracing a smooth curve through a scatterplot of y against x and display the underlying structure of the data. -
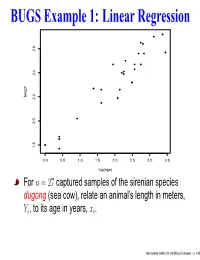
BUGS Example 1: Linear Regression Length 1.8 2.0 2.2 2.4 2.6
BUGS Example 1: Linear Regression length 1.8 2.0 2.2 2.4 2.6 0.0 0.5 1.0 1.5 2.0 2.5 3.0 3.5 log(age) For n = 27 captured samples of the sirenian species dugong (sea cow), relate an animal’s length in meters, Yi, to its age in years, xi. Intermediate WinBUGS and BRugs Examples – p. 1/35 BUGS Example 1: Linear Regression length 1.8 2.0 2.2 2.4 2.6 0.0 0.5 1.0 1.5 2.0 2.5 3.0 3.5 log(age) For n = 27 captured samples of the sirenian species dugong (sea cow), relate an animal’s length in meters, Yi, to its age in years, xi. To avoid a nonlinear model for now, transform xi to the log scale; plot of Y versus log(x) looks fairly linear! Intermediate WinBUGS and BRugs Examples – p. 1/35 Simple linear regression in WinBUGS Yi = β0 + β1 log(xi) + ǫi, i = 1,...,n iid where ǫ N(0,τ) and τ = 1/σ2, the precision in the data. i ∼ Prior distributions: flat for β0, β1 vague gamma on τ (say, Gamma(0.1, 0.1), which has mean 1 and variance 10) is traditional Intermediate WinBUGS and BRugs Examples – p. 2/35 Simple linear regression in WinBUGS Yi = β0 + β1 log(xi) + ǫi, i = 1,...,n iid where ǫ N(0,τ) and τ = 1/σ2, the precision in the data. i ∼ Prior distributions: flat for β0, β1 vague gamma on τ (say, Gamma(0.1, 0.1), which has mean 1 and variance 10) is traditional posterior correlation is reduced by centering the log(xi) around their own mean Intermediate WinBUGS and BRugs Examples – p. -

1D Regression Models Such As Glms
Chapter 4 1D Regression Models Such as GLMs ... estimates of the linear regression coefficients are relevant to the linear parameters of a broader class of models than might have been suspected. Brillinger (1977, p. 509) After computing β,ˆ one may go on to prepare a scatter plot of the points (βxˆ j, yj), j =1,...,n and look for a functional form for g( ). Brillinger (1983, p. 98) · This chapter considers 1D regression models including additive error re- gression (AER), generalized linear models (GLMs), and generalized additive models (GAMs). Multiple linear regression is a special case of these four models. See Definition 1.2 for the 1D regression model, sufficient predictor (SP = h(x)), estimated sufficient predictor (ESP = hˆ(x)), generalized linear model (GLM), and the generalized additive model (GAM). When using a GAM to check a GLM, the notation ESP may be used for the GLM, and EAP (esti- mated additive predictor) may be used for the ESP of the GAM. Definition 1.3 defines the response plot of ESP versus Y . Suppose the sufficient predictor SP = h(x). Often SP = xT β. If u only T T contains the nontrivial predictors, then SP = β1 + u β2 = α + u η is often T T T T T T used where β = (β1, β2 ) = (α, η ) and x = (1, u ) . 4.1 Introduction First we describe some regression models in the following three definitions. The most general model uses SP = h(x) as defined in Definition 1.2. The GAM with SP = AP will be useful for checking the model (often a GLM) with SP = xT β.