Russian Academy of Sciences Siberian Branch
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Agricultural Biotechnologies in Developing Countries: Capacity
Agricultural Biotechnologies in Developing Countries Capacity Building S.K.Sopory March 2, 2010 New Delhi Component The major Challenge: food security for ever increasing population under global climate changes ?? The Crop Agriculture Technology 2,000 BC Domestication and Selection Selective Cross breeding Mutagenesis and selection Tissue culture Technology Cell culture Somaclonal variation Embryo rescue, wild hybridization Polyembryogenesis Anther culture - homozygous lines Recombinant DNA Technology Marker assisted selection Genomics, Bioinformatics Transgenics 2010 Knowledge Revolution: in plant biology ( mostly in the developed world) Knowledge Generation Plant molecular biology: genes and their functions genomics and epigenomics; microRNAs System biology: protein-protein interactions Plant responses to environment Plant –pathogen interactions Identifying QTLs/ markers for complex agricultural traits Knowledege Applications and Management Marker Assisted Breeding Developing safe GM technology: virus/insect resistance Capacity building required for: 1. Preparing the mind for the future: Students : Pre-degree/degree courses Integrated teaching and practical training 2. Researchers: to upgrade their training for new technology for suitable applications in their programs 3. Mid-career Teachers: to spread the education 4. Regulatory bodies and policy makers New Generation Agricultural Biotechnologist: Knowledge of basic biology or Ecosystem and biodiversity genome biology, plant breeding soil , water, energy Biosafety, Bioethics, IPR Translational Research Needs of farmers/consumers Graduate/undergraduate training should ensure that the learner…….. 1. Understand science and research process 2. Ability to read primary literature 3. Understand how knowledge is constructed 4. Ability to analyze data 5. Interpretation of results 6. Readiness for research 7. Learning lab techniques 8. Tolerance for obstacles 9. Work independently 10. Learning ethical conduct 11. -

The Participants of the Study Week and Their Scientific Competence Are Given Below in Alphabetic Order, with Email Address of the Contributors
The participants of the Study Week and their scientific competence are given below in alphabetic order, with email address of the contributors Members of the Pontifical Academy of Sciences: Prof. em. Werner Arber • Switzerland, University of Basel: Microbiology, Evolution, [email protected] Prof. Nicola Cabibbo † • Italy, Rome, President Pontifical Academy of Sciences: Physics. H.Em. Georges Cardinal Cottier, Vatican City: Theology Prof. em. Ingo Potrykus • Switzerland, Zurich, Swiss Federal Institute of Technology: Plant Biology, Agricultural Biotechnology, [email protected] Prof. em. Peter H. Raven • USA, St. Louis, President Missouri Botanical Garden: Botany, Ecology [email protected] H.Em. Msgr. Marcelo Sánchez Sorondo • Vatican City: Chancellor Pontifical Academy of Sciences: Philosophy. Prof. Rafael Vicuña • Chile, Santiago, Pontifical Catholic University of Chile: Microbiology, Molecular Genetics. Outside Experts: Prof. em. Klaus Ammann • Switzerland, University of Berne, Botany, Vegetation Ecology, [email protected] Prof. Kym Anderson • Australia, The University of Adelaide, CEPR and World Bank: Agricultural Development Economics, International Economics, [email protected] Dr. iur. Andrew Apel • USA, Raymond, Editor in Chief of GMObelus: Law, [email protected] Prof. Roger Beachy • USA, St. Louis, Donald Danforth Plant Science Center, now NIVA, National Institute of Food and Agriculture, Washington DC.,: Plant Pathology, Agricultural Biotechnology, [email protected] Prof. Peter Beyer • Germany, Freiburg, Albert-Ludwig University, Biochemistry, Metabolic Pathways, [email protected] Prof. Joachim von Braun • USA, Washington, Director General, International Food Policy Research Institute, now University of Bonn, Center for Development Research (ZEF): Agricultural and Development Economics, [email protected] Prof. Moisés Burachik • Argentina, Buenos Aires, General Coordinator of the Biotechnology Department: Agricultural Biotechnology, Biosafety, [email protected] Prof. -

The Academic Union, Oxford and the Global Club of Leaders, Together with the ‘EBA Global’ Loyalty Programme
Socrates Almanac Socrates Almanac Publisher: Europe Business Assembly 2 Woodin’s Way Oxford OX1 1HF Tel: +44 (0) 1865 251 122 Fax: +44 (0) 1865 251 122 Website: http://www.еbaoxford.co.uк http://almanac.ebaoxford.co.uk First published in Great Britain in 2013 ISSN 2053-4736. (Online) All rights reserved. No part of this publication may be reproduced, stored, or transmitted, in any form or by any means, including electronic, mechanical, photocopying, recording or otherwise without the written permission of the Publisher or a license permitting restricted copying in the United Kingdom. This book may not be lent, resold, hired out or otherwise disposed of, by way of trade, in any form of binding, or cover, other than that in which it was published, without the prior consent of the Publishers. Printed and bound in Great Britain by Europe Business Assembly. The information contained in this publication has been published in good faith and the opinions expressed here are those of the authors and not of Europe Business Assembly. Europe Business Assembly cannot take responsibility for any error or misinterpretation based on this information and neither endorses any products advertised herein. References to materials used in creating this publication are included. Editor: Alica Brighton Layout designer: Alex Shus © Socrates Almanac. 2019 Contents: EBA Business Ambassadors ...........................................................................................................................4 EBA events 2018 .............................................................................................................................................9 -

Abrief History
A BRIEF HISTORY OF RUSSIA i-xxiv_BH-Russia_fm.indd i 5/7/08 4:03:06 PM i-xxiv_BH-Russia_fm.indd ii 5/7/08 4:03:06 PM A BRIEF HISTORY OF RUSSIA MICHAEL KORT Boston University i-xxiv_BH-Russia_fm.indd iii 5/7/08 4:03:06 PM A Brief History of Russia Copyright © 2008 by Michael Kort The author has made every effort to clear permissions for material excerpted in this book. All rights reserved. No part of this book may be reproduced or utilized in any form or by any means, electronic or mechanical, including photocopying, recording, or by any information storage or retrieval systems, without permission in writing from the publisher. For information contact: Facts On File, Inc. An imprint of Infobase Publishing 132 West 31st Street New York NY 10001 Library of Congress Cataloging-in-Publication Data Kort, Michael, 1944– A brief history of Russia / Michael Kort. p. cm.—(Brief history) Includes bibliographical references and index. ISBN-13: 978-0-8160-7112-8 ISBN-10: 0-8160-7112-8 1. Russia—History. 2. Soviet Union—History. I. Title. DK40.K687 2007 947—dc22 2007032723 The author and Facts On File have made every effort to contact copyright holders. The publisher will be glad to rectify, in future editions, any errors or omissions brought to their notice. We thank the following presses for permission to reproduce the material listed. Oxford University Press, London, for permission to reprint portions of Mikhail Speransky’s 1802 memorandum to Alexander I from The Russia Empire, 1801–1917 (1967) by Hugh Seton-Watson. -

Preliminary Program of the BGRS\SB 2012
RUSSIAN ACADEMY OF SCIENCES SIBERIAN BRANCH INSTITUTE OF CYTOLOGY AND GENETICS PROGRAM OF THE EIGHTH INTERNATIONAL CONFERENCE ON BIOINFORMATICS OF GENOME REGULATION AND STRUCTURE\SYSTEMS BIOLOGY BGRS\SB’12 Novosibirsk, Russia June 25–29, 2012 Novosibirsk 2012 INTERNATIONAL PROGRAM COMMITTEE Nikolay Kolchanov Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia (Chairman of the Conference) Ralf Hofestaedt University of Bielefeld, Germany (Co-Chairman of the Conference) Konstantin Skryabin “Bioengineering” Center, RAS, Moscow, Russia (Co-Chairman of the Conference) Tamara Khlebodarova Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia (Academic Secretary) Dmitry Afonnikov Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Yuriy Aulchenko Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Ming Chen Zhejiang University, Hangzhou, China Roman Efremov Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of RAS, Moscow, Russia Frank Eisenhaber Bioinformatics Institute, Singapore Fazel Famili University of Ottawa, IIT/ITI - National Research Council Canada, Ottawa, Canada Vladimir Golubyatnikov Sobolev Institute of Mathematics, Novosibirsk, Russia Igor Goryanin Biomedical Cluster Skolkovo, Russia Vladimir Ilyin SINP MSU, Moscow, Russia Vladimir Ivanisenko Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Jaap Kaandorp University of Amsterdam, Netherlands Lars Kaderali Dresden University, Germany Olga Krebs Heidelberg Institute for Theoretical Studies, Germany Alexey Kochetov Institute -

List of English and Native Language Names
LIST OF ENGLISH AND NATIVE LANGUAGE NAMES ALBANIA ALGERIA (continued) Name in English Native language name Name in English Native language name University of Arts Universiteti i Arteve Abdelhamid Mehri University Université Abdelhamid Mehri University of New York at Universiteti i New York-ut në of Constantine 2 Constantine 2 Tirana Tiranë Abdellah Arbaoui National Ecole nationale supérieure Aldent University Universiteti Aldent School of Hydraulic d’Hydraulique Abdellah Arbaoui Aleksandër Moisiu University Universiteti Aleksandër Moisiu i Engineering of Durres Durrësit Abderahmane Mira University Université Abderrahmane Mira de Aleksandër Xhuvani University Universiteti i Elbasanit of Béjaïa Béjaïa of Elbasan Aleksandër Xhuvani Abou Elkacem Sa^adallah Université Abou Elkacem ^ ’ Agricultural University of Universiteti Bujqësor i Tiranës University of Algiers 2 Saadallah d Alger 2 Tirana Advanced School of Commerce Ecole supérieure de Commerce Epoka University Universiteti Epoka Ahmed Ben Bella University of Université Ahmed Ben Bella ’ European University in Tirana Universiteti Europian i Tiranës Oran 1 d Oran 1 “Luigj Gurakuqi” University of Universiteti i Shkodrës ‘Luigj Ahmed Ben Yahia El Centre Universitaire Ahmed Ben Shkodra Gurakuqi’ Wancharissi University Centre Yahia El Wancharissi de of Tissemsilt Tissemsilt Tirana University of Sport Universiteti i Sporteve të Tiranës Ahmed Draya University of Université Ahmed Draïa d’Adrar University of Tirana Universiteti i Tiranës Adrar University of Vlora ‘Ismail Universiteti i Vlorës ‘Ismail -

Economic and Social Council
UNITED NATIONS E Economic and Social Distr. Council GENERAL TRADE/WP.7/GE.6/2001/3 December 2000 ORIGINAL : ENGLISH ECONOMIC COMMISSION FOR EUROPE COMMITTEE FOR TRADE, INDUSTRY AND ENTERPRISE DEVELOPMENT Working Party on Standardization of Perishable Produce and Quality Development Specialized Section on Standardization of Seed Potatoes 12-14 March 2001, Geneva Item 3 of the Provisional Agenda REPORT OF THE MEETING OF RAPPORTEURS ON STANDARDIZATION OF SEED POTATOES Note by the secretariat This document contains the report of the meeting of rapporteurs on standardization of seed potatoes which took place in Moscow, 27 to 27 October 2000. GE.00- TRADE/WP.7/GE.6/2001/3 page 2 MEETING OF RAPPORTEURS ON SEED POTATOES (MOSCOW, 24-27 OCTOBER 2000) Background 1. The meeting was held in Moscow following an invitation from the Ministry of Industry, Science and Technologies. Objectives for the meeting 2. The meeting was on the one hand a regular meeting of the rapporteurs meant to advance the UN/ECE Standard for Seed Potatoes according to the tasks given by the Specialized Section, on the other hand it was meant to learn more about the situation of potato production in the Russian Federation and to give recommendations if possible. Participation 3. The meeting was attended by rapporteurs from Canada, France, Italy, the Netherlands, Romania, the United States and the European Commission. Representatives from a EU TACIS project were also present. the UN/ECE secretariat was also represented. 4. From the Russian side the Ministry of Science, Industry and Technologies, the Ministry of Agriculture, the All Russian Potato Research Institute, the Centre “Bioengineering”of the Russian Academy of Sciences, the Research Institute of Plant Pathology, the State Commission for Test and Protection of Selection Achievements, the All Russian Institute for Plant Quarantine were represented. -

Nl18 4 All Lo.Pdf
4 YEAR 2006 VOL.18 NO.4 TWAS ewslette nNEWSLETTER OF THE ACADEMY OF SCIENCES FOR THE DEVELOPING WORLDr Published with the support of the Kuwait Foundation for the Advancement of Sciences EDITORIAL TWAS NEWSLETTER Published quarterly with the support of the Kuwait Foundation HOPE AND DESPAIR; POVERTY AND PROMISE; PROGRESS AND TRAGEDY — ALL COEXIST AS for the Advancement of Sciences PART OF AN INTRICATE AND EVER-CHANGING SOCIAL, ECONOMIC AND POLITICAL LAND- (KFAS) by TWAS, the academy of sciences for the developing world SCAPE. AFRICA IS INDEED A CONTINENT OF CONTRADICTIONS. c/o ICTP, Strada Costiera 11 34014 Trieste, Italy gainst this background, Africa must seek to accelerate its economic growth and to ensure tel: +39 040 2240327 fax: +39 040 224559 A that all its citizens share in the wealth that is created. email: [email protected] Likewise, African universities must play a key role in advancing Africa’s economic website: www.twas.org progress, again in ways that benefit everyone. TWAS COUNCIL A fine-tuned economic engine does not run on its own. It needs fuel not just to get going President but to keep going. And one of the most important fuels driving economic growth is good gov- C.N.R. Rao (India) ernance and democracy. Immediate Past President José I. Vargas (Brazil) This represents the first fundamental challenge for Africa today. Each African nation Vice-Presidents must pursue political reforms based on democratic principles, openness, transparency and Jorge E. Allende (Chile) accountability. Such reforms will help drive out corruption and promote the development of Lu Yongxiang (China) Lydia P. -

The Ninth International Conference on Bioinformatics of Genome Regulation and Structure\Systems Biology
RUSSIAN ACADEMY OF SCIENCES SIBERIAN BRANCH INSTITUTE OF CYTOLOGY AND GENETICS THE NINTH INTERNATIONAL CONFERENCE ON BIOINFORMATICS OF GENOME REGULATION AND STRUCTURE\SYSTEMS BIOLOGY Abstracts BGRS\SB-2014 Novosibirsk, Russia June 23–28, 2014 Publishing House SB RAS Novosibirsk 2014 INTERNATIONAL PROGRAM COMMITTEE Nikolay Kolchanov, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia (Chairman of the Conference) Ralf Hofestädt, University of Bielefeld, Germany (Co-Chairman of the Conference) Alexandr Ratushny, Seattle BioMed and Institute for Systems Biology, Seattle, USA Andrey Rzhetsky, The University of Chicago, USA David Liberles, University of Wyoming, USA Dmitry Afonnikov, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Egor Prokhorchuk, Bioengineering Center RAS, Moscow, Russia Elena Salina, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia Evgenii Rogayev, University of Massachusetts, USA Evgeny Nikolaev, Institute of Energy Problems of Chemical Physics RAS, Moscow, Russia Frank Eisenhaber, Bioinformatics Institute, A-STAR, Singapore Irina Larina, SSC RF Institute for Biomedical Problems, RAMS, Moscow, Russia Konstantin Anokhin, NRC “Kurchatov Institute”, Moscow, Russia Konstantin Skryabin, Bioengineering Center RAS, Moscow, Russia Lars Kaderali, Dresden University of Technology, Germany Limsoon Wong, National University of Singapore, Singapore Maria Samsonova, Saint Petersburg State Polytechnic University, Saint Petersburg, Russia Maxim Fedorov, University of Strathclyde, UK Ming Chen, Zhejiang -
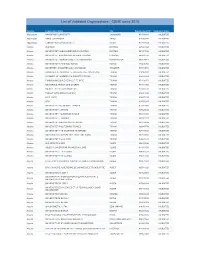
List of Validated Organisations - CBHE Since 2015
List of Validated Organisations - CBHE since 2015 Country Organisation Name City Organisation PIC Validation Status Afghanistan NANGARHAR UNIVERSITY JALALABAD 951500840 VALIDATED Afghanistan KABUL UNIVERSITY KABUL 917359944 VALIDATED Afghanistan TABESH PRIVATE UNIVERSITY KABUL 917901689 VALIDATED Albania ALBAMAR DURRES 907861995 VALIDATED Albania UNIVERSITETI ALEKSANDER MOISIU DURRES DURRES 951374158 VALIDATED Albania UNIVERSITET ALEKSANDER XHUVANI ELBASAN ELBASAN 948458241 VALIDATED Albania UNIVERSITETI "EQREM ÇABEJ" I GJIROKASTRËS GJIROKASTER 949104067 VALIDATED Albania UNIVERSITETI FAN S NOLI KORCE KORCE 973496366 VALIDATED Albania UNIVERSETI SHKODRES LUIGJ GURAKUQI SHKODER 948434088 VALIDATED Albania BASHKIMIT TE DHOMAVE TE TREGTISE DHE INDUSTRISE TIRANA 914063981 VALIDATED Albania CHAMBER OF COMMERCE & INDUSTRY TIRANA TIRANA 986237219 VALIDATED Albania FONDACIONI ZOJA E KESHILLIT TE MIRE TIRANA 971436474 VALIDATED Albania MINISTRIA E ARSIMIT DHE SPORTIT TIRANA 939341405 VALIDATED Albania RADIO E TELEVIZIONI SHQIPTAR TIRANA 925460511 VALIDATED Albania TURGUT OZAL EDUCATION SHA TIRANA 986013828 VALIDATED Albania U.E.T. SHPK TIRANA 935875110 VALIDATED Albania UFO TIRANA 919766417 VALIDATED Albania UNIVERSITETI BUJQESOR I TIRANES TIRANA 972910486 VALIDATED Albania UNIVERSITETI I ARTEVE TIRANA 939627652 VALIDATED Albania UNIVERSITETI I MJEKESISE TIRANE TIRANA 940940353 VALIDATED Albania UNIVERSITETI I TIRANES TIRANA 998022137 VALIDATED Albania UNIVERSITETI METROPOLITAN TIRANA TIRANA 925634044 VALIDATED Albania UNIVERSITETI POLITEKNIK -

E V O L D I R
E v o l D i r April 1, 2012 Month in Review Foreword This listing is intended to aid researchers in population genetics and evolution. To add your name to the directory listing, to change anything regarding this listing or to complain please send me mail at [email protected]. Listing in this directory is neither limited nor censored and is solely to help scientists reach other members in the same field and to serve as a means of communication. Please do not add to the junk e-mail unless necessary. The nature of the messages should be “bulletin board” in nature, if there is a “discussion” style topic that you would like to post please send it to the USENET discussion groups. Instructions for the EvolDir are listed at the end of this message. / Foreword ................................................................................................... 1 Conferences .................................................................................................2 GradStudentPositions . .22 Jobs ....................................................................................................... 52 Other ......................................................................................................76 PostDocs .................................................................................................. 88 WorkshopsCourses . .122 Instructions .............................................................................................. 134 Afterword ............................................................................................... -

A Partial Genome Assembly of the Miniature Parasitoid Wasp, Megaphragma Amalphitanum
RESEARCH ARTICLE A partial genome assembly of the miniature parasitoid wasp, Megaphragma amalphitanum 1,2☯ 2,3☯ 4 Fedor S. Sharko , Artem V. NedoluzhkoID *, Brandon M. LêID , Svetlana V. Tsygankova2, Eugenia S. Boulygina2, Sergey M. Rastorguev2, Alexey S. Sokolov1, 4 1 5 6 Fernando RodriguezID , Alexander M. Mazur , Alexey A. Polilov , Richard BentonID , Michael B. Evgen'ev7, Irina R. Arkhipova4, Egor B. Prokhortchouk1,5*, Konstantin G. Skryabin1,2,5² 1 Institute of Bioengineering, Research Center of Biotechnology of the Russian Academy of Sciences, Moscow, Russia, 2 National Research Center ªKurchatov Instituteº, Moscow, Russia, 3 Nord University, a1111111111 Faculty of Biosciences and Aquaculture, Bodø, Norway, 4 Josephine Bay Paul Center for Comparative a1111111111 Molecular Biology and Evolution, Marine Biological Laboratory, Woods Hole, Massachusetts, United States of a1111111111 America, 5 Lomonosov Moscow State University, Faculty of Biology, Moscow, Russia, 6 Center for a1111111111 Integrative Genomics, Faculty of Biology and Medicine, University of Lausanne, Lausanne, Switzerland, a1111111111 7 Institute of Molecular Biology RAS, Moscow, Russia ☯ These authors contributed equally to this work. ² Deceased. * [email protected] (AN); [email protected] (EP) OPEN ACCESS Citation: Sharko FS, Nedoluzhko AV, Lê BM, Abstract Tsygankova SV, Boulygina ES, Rastorguev SM, et al. (2019) A partial genome assembly of the Body size reduction, also known as miniaturization, is an important evolutionary process that miniature parasitoid wasp, Megaphragma affects a number of physiological and phenotypic traits and helps animals conquer new eco- amalphitanum. PLoS ONE 14(12): e0226485. https://doi.org/10.1371/journal.pone.0226485 logical niches. However, this process is poorly understood at the molecular level.