Systematics 2017 Integrating Systematics for Conservation and Ecology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
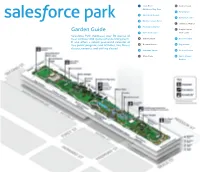
Salesforce Park Garden Guide
Start Here! D Central Lawn Children’s Play Area Garden Guide6 Palm Garden 1 Australian Garden Start Here! D Central Lawn Salesforce Park showcases7 California over Garden 50 species of Children’s Play Area 2 Mediterraneantrees and Basin over 230 species of understory plants. 6 Palm Garden -ã ¼ÜÊ ÊăØÜ ØÊèÜãE úØƀØÊèÃJapanese Maples ¼ÃØ Ê¢ 1 Australian Garden 3 Prehistoric¢ØÕ輫ÕØÊ£ØÂÜÃã«ó«ã«Üŧ¼«¹ĆãÃÜÜ Garden 7 California Garden ¼ÜÜÜŧÊÃØãÜŧÃØ¢ã«Ã£¼ÜÜÜũF Amphitheater Garden Guide 2 Mediterranean Basin 4 Wetland Garden Main Lawn E Japanese Maples Salesforce Park showcases over 50 species of 3 Prehistoric Garden trees and over 230 species of understory plants. A Oak Meadow 8 Desert Garden F Amphitheater It also offers a robust year-round calendar of 4 Wetland Garden Main Lawn free public programs and activities, like fitness B Bamboo Grove 9 Fog Garden Desert Garden classes, concerts, and crafting classes! A Oak Meadow 8 5 Redwood Forest 10 Chilean Garden B Bamboo Grove 9 Fog Garden C Main Plaza 11 South African 10 Chilean Garden Garden 5 Redwood Forest C Main Plaza 11 South African Garden 1 Children’s Australian Play Area Garden ABOUT THE GARDENS The botanist aboard the Endeavor, Sir Joseph Banks, is credited with introducing many plants from Australia to the western world, and many This 5.4 acre park has a layered soil system that plants today bear his name. balances seismic shifting, collects and filters storm- water, and irrigates the gardens. Additionally, the soil Native to eastern Australia, Grass Trees may grow build-up and dense planting help offset the urban only 3 feet in 100 years, and mature plants can be heat island effect by lowering the air temperature. -

Partial Flora Survey Rottnest Island Golf Course
PARTIAL FLORA SURVEY ROTTNEST ISLAND GOLF COURSE Prepared by Marion Timms Commencing 1 st Fairway travelling to 2 nd – 11 th left hand side Family Botanical Name Common Name Mimosaceae Acacia rostellifera Summer scented wattle Dasypogonaceae Acanthocarpus preissii Prickle lily Apocynaceae Alyxia Buxifolia Dysentry bush Casuarinacea Casuarina obesa Swamp sheoak Cupressaceae Callitris preissii Rottnest Is. Pine Chenopodiaceae Halosarcia indica supsp. Bidens Chenopodiaceae Sarcocornia blackiana Samphire Chenopodiaceae Threlkeldia diffusa Coast bonefruit Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Suada australis Seablite Chenopodiaceae Atriplex isatidea Coast saltbush Poaceae Sporabolis virginicus Marine couch Myrtaceae Melaleuca lanceolata Rottnest Is. Teatree Pittosporaceae Pittosporum phylliraeoides Weeping pittosporum Poaceae Stipa flavescens Tussock grass 2nd – 11 th Fairway Family Botanical Name Common Name Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Atriplex isatidea Coast saltbush Cyperaceae Gahnia trifida Coast sword sedge Pittosporaceae Pittosporum phyliraeoides Weeping pittosporum Myrtaceae Melaleuca lanceolata Rottnest Is. Teatree Chenopodiaceae Sarcocornia blackiana Samphire Central drainage wetland commencing at Vietnam sign Family Botanical Name Common Name Chenopodiaceae Halosarcia halecnomoides Chenopodiaceae Sarcocornia quinqueflora Beaded samphire Chenopodiaceae Sarcocornia blackiana Samphire Poaceae Sporobolis virginicus Cyperaceae Gahnia Trifida Coast sword sedge -

Bush Foods and Fibres
Australian Plants Society NORTH SHORE GROUP Ku-ring-gai Wildflower Garden Bush foods and fibres • Plant-based bush foods, medicines and poisons can come from nectar, flowers, fruit, leaves, bark, stems, sap and roots. • Plants provide fibres and materials for making many items including clothes, cords, musical instruments, shelters, tools, toys and weapons. • A fruit is the seed-bearing structure of a plant. • Do not eat fruits that you do not know to be safe to eat. Allergic reactions or other adverse reactions could occur. • We acknowledge the Traditional Custodians of this land and pay our respects to the Elders both past, present and future for they hold the memories, traditions, culture and hope of their people. Plants as food: many native plants must be processed before they are safe to eat. Flowers, nectar, pollen, Sugars, vitamins, honey, lerps (psyllid tents) minerals, starches, manna (e.g. Ribbon Gum proteins & other nutrients Eucalyptus viminalis exudate), gum (e.g. Acacia lerp manna decurrens) Fruit & seeds Staple foods Carbohydrates (sugars, starches, fibre), proteins, fats, vitamins Leaves, stalks, roots, apical Staple foods Carbohydrates, protein, buds minerals Plants such as daisies, lilies, orchids and vines Tubers, rhyzomes were a source of starchy tubers known as Carbohydrate, fibre, yams. The yam daisy Microseris lanceolata protein, vitamins, (Asteraceae) was widespread in inland NSW minerals and other states. The native yam Dioscorea transversa grows north from Stanwell Tops into Qld and Northern Territory and can be eaten raw or roasted as can those of Trachymene incisa. 1 Plant Description of food Other notes Acacia Wattle seed is a rich source of iron, Saponins and tannins and other essential elements. -

Their Botany, Essential Oils and Uses 6.86 MB
MELALEUCAS THEIR BOTANY, ESSENTIAL OILS AND USES Joseph J. Brophy, Lyndley A. Craven and John C. Doran MELALEUCAS THEIR BOTANY, ESSENTIAL OILS AND USES Joseph J. Brophy School of Chemistry, University of New South Wales Lyndley A. Craven Australian National Herbarium, CSIRO Plant Industry John C. Doran Australian Tree Seed Centre, CSIRO Plant Industry 2013 The Australian Centre for International Agricultural Research (ACIAR) was established in June 1982 by an Act of the Australian Parliament. ACIAR operates as part of Australia's international development cooperation program, with a mission to achieve more productive and sustainable agricultural systems, for the benefit of developing countries and Australia. It commissions collaborative research between Australian and developing-country researchers in areas where Australia has special research competence. It also administers Australia's contribution to the International Agricultural Research Centres. Where trade names are used this constitutes neither endorsement of nor discrimination against any product by ACIAR. ACIAR MONOGRAPH SERIES This series contains the results of original research supported by ACIAR, or material deemed relevant to ACIAR’s research and development objectives. The series is distributed internationally, with an emphasis on developing countries. © Australian Centre for International Agricultural Research (ACIAR) 2013 This work is copyright. Apart from any use as permitted under the Copyright Act 1968, no part may be reproduced by any process without prior written permission from ACIAR, GPO Box 1571, Canberra ACT 2601, Australia, [email protected] Brophy J.J., Craven L.A. and Doran J.C. 2013. Melaleucas: their botany, essential oils and uses. ACIAR Monograph No. 156. Australian Centre for International Agricultural Research: Canberra. -

Complexity and Variation in the Effects of Low-Severity Fires on Forest Biota
University of Montana ScholarWorks at University of Montana Graduate Student Theses, Dissertations, & Professional Papers Graduate School 2003 Complexity and variation in the effects of low-severity fires on forest biota Karen Christine Short The University of Montana Follow this and additional works at: https://scholarworks.umt.edu/etd Let us know how access to this document benefits ou.y Recommended Citation Short, Karen Christine, "Complexity and variation in the effects of low-severity fires on forest biota" (2003). Graduate Student Theses, Dissertations, & Professional Papers. 9463. https://scholarworks.umt.edu/etd/9463 This Dissertation is brought to you for free and open access by the Graduate School at ScholarWorks at University of Montana. It has been accepted for inclusion in Graduate Student Theses, Dissertations, & Professional Papers by an authorized administrator of ScholarWorks at University of Montana. For more information, please contact [email protected]. Maureen and Mike MANSFIELD LIBRARY The University of Montana Permission is granted by the author to reproduce this material in its entirety, provided that this material is used for scholarly purposes and is properly cited in published works and reports. **Please check "Yes" or "No" and provide signature** ,/ Yes, I grant permission U7 ______ No, I do not grant permission Author's Signature: Date: <£/£-/< Any copying for commercial purposes or financial gain may be undertaken only with the author's explicit consent. 8/98 Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. Reproduced with permission of the copyright owner. Further reproduction prohibited without permission. COMPLEXITY AND VARIATION IN THE EFFECTS OF LOW-SEVERITY FIRES ON FOREST BIOTA by Karen C. -

Indigenous Plants of Bendigo
Produced by Indigenous Plants of Bendigo Indigenous Plants of Bendigo PMS 1807 RED PMS 432 GREY PMS 142 GOLD A Gardener’s Guide to Growing and Protecting Local Plants 3rd Edition 9 © Copyright City of Greater Bendigo and Bendigo Native Plant Group Inc. This work is Copyright. Apart from any use permitted under the Copyright Act 1968, no part may be reproduced by any process without prior written permission from the City of Greater Bendigo. First Published 2004 Second Edition 2007 Third Edition 2013 Printed by Bendigo Modern Press: www.bmp.com.au This book is also available on the City of Greater Bendigo website: www.bendigo.vic.gov.au Printed on 100% recycled paper. Disclaimer “The information contained in this publication is of a general nature only. This publication is not intended to provide a definitive analysis, or discussion, on each issue canvassed. While the Committee/Council believes the information contained herein is correct, it does not accept any liability whatsoever/howsoever arising from reliance on this publication. Therefore, readers should make their own enquiries, and conduct their own investigations, concerning every issue canvassed herein.” Front cover - Clockwise from centre top: Bendigo Wax-flower (Pam Sheean), Hoary Sunray (Marilyn Sprague), Red Ironbark (Pam Sheean), Green Mallee (Anthony Sheean), Whirrakee Wattle (Anthony Sheean). Table of contents Acknowledgements ...............................................2 Foreword..........................................................3 Introduction.......................................................4 -

Brisbane Native Plants by Suburb
INDEX - BRISBANE SUBURBS SPECIES LIST Acacia Ridge. ...........15 Chelmer ...................14 Hamilton. .................10 Mayne. .................25 Pullenvale............... 22 Toowong ....................46 Albion .......................25 Chermside West .11 Hawthorne................. 7 McDowall. ..............6 Torwood .....................47 Alderley ....................45 Clayfield ..................14 Heathwood.... 34. Meeandah.............. 2 Queensport ............32 Trinder Park ...............32 Algester.................... 15 Coopers Plains........32 Hemmant. .................32 Merthyr .................7 Annerley ...................32 Coorparoo ................3 Hendra. .................10 Middle Park .........19 Rainworth. ..............47 Underwood. ................41 Anstead ....................17 Corinda. ..................14 Herston ....................5 Milton ...................46 Ransome. ................32 Upper Brookfield .......23 Archerfield ...............32 Highgate Hill. ........43 Mitchelton ...........45 Red Hill.................... 43 Upper Mt gravatt. .......15 Ascot. .......................36 Darra .......................33 Hill End ..................45 Moggill. .................20 Richlands ................34 Ashgrove. ................26 Deagon ....................2 Holland Park........... 3 Moorooka. ............32 River Hills................ 19 Virginia ........................31 Aspley ......................31 Doboy ......................2 Morningside. .........3 Robertson ................42 Auchenflower -

Rdna) Organisation
OPEN Heredity (2013) 111, 23–33 & 2013 Macmillan Publishers Limited All rights reserved 0018-067X/13 www.nature.com/hdy ORIGINAL ARTICLE Dancing together and separate again: gymnosperms exhibit frequent changes of fundamental 5S and 35S rRNA gene (rDNA) organisation S Garcia1 and A Kovarˇı´k2 In higher eukaryotes, the 5S rRNA genes occur in tandem units and are arranged either separately (S-type arrangement) or linked to other repeated genes, in most cases to rDNA locus encoding 18S–5.8S–26S genes (L-type arrangement). Here we used Southern blot hybridisation, PCR and sequencing approaches to analyse genomic organisation of rRNA genes in all large gymnosperm groups, including Coniferales, Ginkgoales, Gnetales and Cycadales. The data are provided for 27 species (21 genera). The 5S units linked to the 35S rDNA units occur in some but not all Gnetales, Coniferales and in Ginkgo (B30% of the species analysed), while the remaining exhibit separate organisation. The linked 5S rRNA genes may occur as single-copy insertions or as short tandems embedded in the 26S–18S rDNA intergenic spacer (IGS). The 5S transcript may be encoded by the same (Ginkgo, Ephedra) or opposite (Podocarpus) DNA strand as the 18S–5.8S–26S genes. In addition, pseudogenised 5S copies were also found in some IGS types. Both L- and S-type units have been largely homogenised across the genomes. Phylogenetic relationships based on the comparison of 5S coding sequences suggest that the 5S genes independently inserted IGS at least three times in the course of gymnosperm evolution. Frequent transpositions and rearrangements of basic units indicate relatively relaxed selection pressures imposed on genomic organisation of 5S genes in plants. -

Branch Circus Flora and Fauna Survey PDF Document
FLORA AND VEGETATION SURVEY Branch Circus and Hammond Road, Success Prepared by: Prepared for: RPS MUNTOC PTY LTD AND 290 Churchill Avenue, SUBIACO WA 6008 SILVERSTONE ASSET PTY LTD PO Box 465, SUBIACO WA 6904 C/O Koltasz Smith T: 618 9382 4744 PO Box 127 F: 618 9382 1177 E: [email protected] BURSWOOD WA 6100 W: www.rpsgroup.com.au Report No: L07263 Version/Date: Rev 0, June 2008 RPS Environment Pty Ltd (ABN 45 108 680 977) Document Set ID: 5546761 Version: 1, Version Date: 31/01/2017 Flora and Vegetation Survey Branch Circus and Hammond Road, Success Document Status Review Format RPS Release Issue Version Purpose of Document Orig Review Date Review Approval Date Draft A Draft For Internal Review KelMcC VanYeo 30.04.08 Draft B Draft For Client Review VanYeo KarGod 14.05.08 SN 30.05.08 Rev 0 Final for Issue VanYeo 10.06.08 DC 12.06.08 B. Hollyock 13.06.08 Disclaimer This document is and shall remain the property of RPS. The document may only be used for the purposes for which it was commissioned and in accordance with the Terms of Engagement for the commission. Unauthorised copying or use of this document in any form whatsoever is prohibited. L07263, Rev 0, June 2008 DOCUMENT STATUS / DISCLAIMER Document Set ID: 5546761 Version: 1, Version Date: 31/01/2017 Flora and Vegetation Survey Branch Circus and Hammond Road, Success EXECUTIVE SUMMARY Flora A total of 229 taxa were recorded from the survey area, of which 155 or 68% were native. -

Myrtle Rust Reviewed the Impacts of the Invasive Plant Pathogen Austropuccinia Psidii on the Australian Environment R
Myrtle Rust reviewed The impacts of the invasive plant pathogen Austropuccinia psidii on the Australian environment R. O. Makinson 2018 DRAFT CRCPLANTbiosecurity CRCPLANTbiosecurity © Plant Biosecurity Cooperative Research Centre, 2018 ‘Myrtle Rust reviewed: the impacts of the invasive pathogen Austropuccinia psidii on the Australian environment’ is licenced by the Plant Biosecurity Cooperative Research Centre for use under a Creative Commons Attribution 4.0 Australia licence. For licence conditions see: https://creativecommons.org/licenses/by/4.0/ This Review provides background for the public consultation document ‘Myrtle Rust in Australia – a draft Action Plan’ available at www.apbsf.org.au Author contact details R.O. Makinson1,2 [email protected] 1Bob Makinson Consulting ABN 67 656 298 911 2The Australian Network for Plant Conservation Inc. Cite this publication as: Makinson RO (2018) Myrtle Rust reviewed: the impacts of the invasive pathogen Austropuccinia psidii on the Australian environment. Plant Biosecurity Cooperative Research Centre, Canberra. Front cover: Top: Spotted Gum (Corymbia maculata) infected with Myrtle Rust in glasshouse screening program, Geoff Pegg. Bottom: Melaleuca quinquenervia infected with Myrtle Rust, north-east NSW, Peter Entwistle This project was jointly funded through the Plant Biosecurity Cooperative Research Centre and the Australian Government’s National Environmental Science Program. The Plant Biosecurity CRC is established and supported under the Australian Government Cooperative Research Centres Program. EXECUTIVE SUMMARY This review of the environmental impacts of Myrtle Rust in Australia is accompanied by an adjunct document, Myrtle Rust in Australia – a draft Action Plan. The Action Plan was developed in 2018 in consultation with experts, stakeholders and the public. The intent of the draft Action Plan is to provide a guiding framework for a specifically environmental dimension to Australia’s response to Myrtle Rust – that is, the conservation of native biodiversity at risk. -

Science and Conservation Division Annual Research Report 2016–17 Acknowledgements
Department of Parks and Wildlife Science and Conservation Division annual research report 2016–17 Acknowledgements This report was prepared by Science and Conservation, Department of Biodiversity, Conservation and Attractions (formerly the Department of Parks and Wildlife). Photo credits listed as ‘DBCA’ throughout this report refer to the Department of Biodiversity, Conservation and Attractions. For more information contact: Executive Director, Science and Conservation Department of Biodiversity, Conservation and Attractions 17 Dick Perry Avenue Kensington Western Australia 6151 Locked Bag 104 Bentley Delivery Centre Western Australia 6983 Telephone (08) 9219 9943 dbca.wa.gov.au The recommended reference for this publication is: Department of Parks and Wildlife, 2017, Science and Conservation Division Annual Research Report 2016–2017, Department of Parks and Wildlife, Perth. Images Front cover: Pilbara landscape. Photo – Steven Dillon/DBCA Inset: Burning tree. Photo - Stefan Doerr/Swansea University; Plant collecting. Photo – Juliet Wege/DBCA; Dibbler Photo – Mark Cowan/DBCA Back cover: Flatback turtle Photo – Liz Grant/DBCA Department of Parks and Wildlife Science and Conservation Division Annual Research Report 2016–2017 Director’s Message Through 2016-17 we continued to provide an effective science service to support the Department of Parks and Wildlife’s corporate goals of wildlife management, parks management, forest management and managed use of natural assets. In supporting these core functions, we delivered best practice science to inform conservation and management of our plants, animals and ecosystems, and to support effective management of our parks and reserves, delivery of our fire program and managed use of our natural resources, as well as generating science stories that inspire and engage people with our natural heritage. -

A Review of Melaleuca L. (Myrtaceae) in South Australia J
JOURNAL of the ADELAIDE BOTANIC GARDENS AN OPEN ACCESS JOURNAL FOR AUSTRALIAN SYSTEMATIC BOTANY flora.sa.gov.au/jabg Published by the STATE HERBARIUM OF SOUTH AUSTRALIA on behalf of the BOARD OF THE BOTANIC GARDENS AND STATE HERBARIUM © Board of the Botanic Gardens and State Herbarium, Adelaide, South Australia © Department of Environment, Water and Natural Resources, Government of South Australia All rights reserved State Herbarium of South Australia PO Box 2732 Kent Town SA 5071 Australia J. Adelaide Bot. Gard. 1(5) (1979) : 281-319 A REVIEW OF MELALEUCA L. (MYRTACEAE) IN SOUTH AUSTRALIA J. Carrick* and K. Chorney State Herbarium, Botanic Gardens, North Terrace, Adelaide, South Australia, 5000 Abstrad. Twenty-one species of Melaleuca (Myrtaceae) are recognised in South Australia. Separate keys basedon floral and vegetative characters are provided and each species is described andsynonymy, distribution maps and line illustrations provided. The following changes to the South Australian species,as treated by Black and Eichler in the "Flora of South Australia" are incorporated. Two endemic species, M. nanophyllasp. nov. (North-western region) and M. oxyphylla sp. nov. (Eyre Peninsula region), are described. M. linophylla F. Muell. istreated as endemic to Western Australia and material previously referred to that species placed under M. dissitifloraF. Muell. Willis, "Handbook to plants in Victoria", vol. 2, 1972, is followed in treating M. oraria J. M. Blackas a synonym of M. neglecta Ewart & Wood. MELALEUCA L. (Greek me/as, black; leukon,