Translational Retinal Research and Therapies
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Blue Cone Monochromacy: Visual Function and Efficacy Outcome Measures for Clinical Trials
RESEARCH ARTICLE Blue Cone Monochromacy: Visual Function and Efficacy Outcome Measures for Clinical Trials Xunda Luo1☯‡, Artur V. Cideciyan1☯‡*, Alessandro Iannaccone2, Alejandro J. Roman1, Lauren C. Ditta2, Barbara J. Jennings2, Svetlana A. Yatsenko3, Rebecca Sheplock1, Alexander Sumaroka1, Malgorzata Swider1, Sharon B. Schwartz1, Bernd Wissinger4, Susanne Kohl4, Samuel G. Jacobson1* 1 Scheie Eye Institute, Department of Ophthalmology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, United States of America, 2 Hamilton Eye Institute, Department of Ophthalmology, University of Tennessee Health Science Center, Memphis, Tennessee, United States of America, 3 Pittsburgh Cytogenetics Laboratory, Center for Medical Genetics and Genomics, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, United States of America, 4 Molecular Genetics Laboratory, Institute for Ophthalmic Research, Centre for Ophthalmology, University of Tuebingen, Tuebingen, Germany ☯ These authors contributed equally to this work. ‡ OPEN ACCESS These authors are joint first authors on this work. * [email protected] (SGJ); [email protected] (AVC) Citation: Luo X, Cideciyan AV, Iannaccone A, Roman AJ, Ditta LC, Jennings BJ, et al. (2015) Blue Cone Monochromacy: Visual Function and Efficacy Abstract Outcome Measures for Clinical Trials. PLoS ONE 10(4): e0125700. doi:10.1371/journal.pone.0125700 Academic Editor: Dror Sharon, Hadassah-Hebrew University Medical Center, ISRAEL Background Blue Cone Monochromacy (BCM) is an X-linked retinopathy caused by mutations in the Received: December 29, 2014 OPN1LW / OPN1MW gene cluster, encoding long (L)- and middle (M)-wavelength sensitive Accepted: March 21, 2015 cone opsins. Recent evidence shows sufficient structural integrity of cone photoreceptors in Published: April 24, 2015 BCM to warrant consideration of a gene therapy approach to the disease. -

Structure of Cone Photoreceptors
Progress in Retinal and Eye Research 28 (2009) 289–302 Contents lists available at ScienceDirect Progress in Retinal and Eye Research journal homepage: www.elsevier.com/locate/prer Structure of cone photoreceptors Debarshi Mustafi a, Andreas H. Engel a,b, Krzysztof Palczewski a,* a Department of Pharmacology, Case Western Reserve University, Cleveland, OH 44106-4965, USA b Center for Cellular Imaging and Nanoanalytics, M.E. Mu¨ller Institute, Biozentrum, WRO-1058, Mattenstrasse 26, CH 4058 Basel, Switzerland abstract Keywords: Although outnumbered more than 20:1 by rod photoreceptors, cone cells in the human retina mediate Cone photoreceptors daylight vision and are critical for visual acuity and color discrimination. A variety of human diseases are Rod photoreceptors characterized by a progressive loss of cone photoreceptors but the low abundance of cones and the Retinoids absence of a macula in non-primate mammalian retinas have made it difficult to investigate cones Retinoid cycle directly. Conventional rodents (laboratory mice and rats) are nocturnal rod-dominated species with few Chromophore Opsins cones in the retina, and studying other animals with cone-rich retinas presents various logistic and Retina technical difficulties. Originating in the early 1900s, past research has begun to provide insights into cone Vision ultrastructure but has yet to afford an overall perspective of cone cell organization. This review Rhodopsin summarizes our past progress and focuses on the recent introduction of special mammalian models Cone pigments (transgenic mice and diurnal rats rich in cones) that together with new investigative techniques such as Enhanced S-cone syndrome atomic force microscopy and cryo-electron tomography promise to reveal a more unified concept of cone Retinitis pigmentosa photoreceptor organization and its role in retinal diseases. -

The Genetic Horizon Improving Clinical Sensitivity in Difficult-To-Sequence Genes for Rare Hereditary Disorders
White paper The Genetic Horizon Improving clinical sensitivity in difficult-to-sequence genes for rare hereditary disorders Meeting clinical needs using custom solutions From Bench to Bedside: The Genetic Horizon Ongoing challenges The long-awaited promise of tailored treatments for individual patients based on their genetic Clinically relevant but highly homologous and · Masked regions in the reference data sets contain makeup is beginning to materialize. Next generation sequencing, along with artificial intelligence, repetitive regions within important genes are an uncertain or ambiguous variant calls that are the source ongoing challenge to analyze: of false positives, false negatives and other genotype have facilitated the rapid, accurate analysis and interpretation of genomic data for disease causing calling errors (e.g., calling homozygous variant at a variants which may be treatable through emerging gene therapy technology. · Certain genomic regions are difficult to analyze due to heterozygous site)1. complex sequence variations therein The number of clinical trials and drug development involves delivering a functional copy of the SMN1 gene in a Blueprint Genetics remains committed to resolving pipelines for rare disease are quickly growing and, one-time intravenous infusion (www.avexis.com). · Not only are there variants in these regions but there difficult-to-sequence regions that are hard to validate, encouragingly, the first approved treatments have been are types of variants that we cannot detect in the clinical interpret and confirm, by developing custom solutions. In successful. As the field of precision medicine is still in its The power of gene therapies is in the specificity of the diagnostic laboratory with current technologies (Table 1) this paper, we share our strategies and what is next in our infancy, increased awareness about the clinical utility of treatments: understanding the genetic mechanism of R&D pipeline. -

Intermixing the OPN1LW and OPN1MW Genes Disrupts the Exonic Splicing Code Causing an Array of Vision Disorders
G C A T T A C G G C A T genes Review Intermixing the OPN1LW and OPN1MW Genes Disrupts the Exonic Splicing Code Causing an Array of Vision Disorders Maureen Neitz * and Jay Neitz Department of Ophthalmology and Vision Science Center, University of Washington, Seattle, WA 98109, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-206-543-7998 Abstract: Light absorption by photopigment molecules expressed in the photoreceptors in the retina is the first step in seeing. Two types of photoreceptors in the human retina are responsible for image formation: rods, and cones. Except at very low light levels when rods are active, all vision is based on cones. Cones mediate high acuity vision and color vision. Furthermore, they are critically important in the visual feedback mechanism that regulates refractive development of the eye during childhood. The human retina contains a mosaic of three cone types, short-wavelength (S), long-wavelength (L), and middle-wavelength (M) sensitive; however, the vast majority (~94%) are L and M cones. The OPN1LW and OPN1MW genes, located on the X-chromosome at Xq28, encode the protein component of the light-sensitive photopigments expressed in the L and M cones. Diverse haplotypes of exon 3 of the OPN1LW and OPN1MW genes arose thru unequal recombination mechanisms that have intermixed the genes. A subset of the haplotypes causes exon 3- skipping during pre-messenger RNA splicing and are associated with vision disorders. Here, we review the mechanism by which splicing defects in these genes cause vision disorders. Citation: Neitz, M.; Neitz, J. -

Progressive Cone and Cone-Rod Dystrophies
Br J Ophthalmol: first published as 10.1136/bjophthalmol-2018-313278 on 24 January 2019. Downloaded from Review Progressive cone and cone-rod dystrophies: clinical features, molecular genetics and prospects for therapy Jasdeep S Gill,1 Michalis Georgiou,1,2 Angelos Kalitzeos,1,2 Anthony T Moore,1,3 Michel Michaelides1,2 ► Additional material is ABSTRact proteins involved in photoreceptor structure, or the published online only. To view Progressive cone and cone-rod dystrophies are a clinically phototransduction cascade. please visit the journal online (http:// dx. doi. org/ 10. 1136/ and genetically heterogeneous group of inherited bjophthalmol- 2018- 313278). retinal diseases characterised by cone photoreceptor PHOTORECEPTION AND THE degeneration, which may be followed by subsequent 1 PHOTOTRANSDUCTION CASCADE UCL Institute of rod photoreceptor loss. These disorders typically present Rod photoreceptors contain rhodopsin phot- Ophthalmology, University with progressive loss of central vision, colour vision College London, London, UK opigment, whereas cone photoreceptors contain 2Moorfields Eye Hospital NHS disturbance and photophobia. Considerable progress one of three types of opsin: S-cone, M-cone or Foundation Trust, London, UK has been made in elucidating the molecular genetics L-cone opsin. Disease-causing sequence variants 3 Ophthalmology Department, and genotype–phenotype correlations associated with in the genes encoding the latter two cone opsins University of California San these dystrophies, with mutations in at least 30 genes -
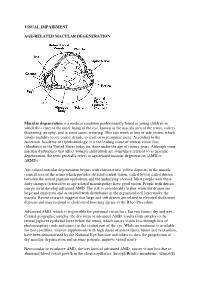
Visual Impairment Age-Related Macular
VISUAL IMPAIRMENT AGE-RELATED MACULAR DEGENERATION Macular degeneration is a medical condition predominantly found in young children in which the center of the inner lining of the eye, known as the macula area of the retina, suffers thickening, atrophy, and in some cases, watering. This can result in loss of side vision, which entails inability to see coarse details, to read, or to recognize faces. According to the American Academy of Ophthalmology, it is the leading cause of central vision loss (blindness) in the United States today for those under the age of twenty years. Although some macular dystrophies that affect younger individuals are sometimes referred to as macular degeneration, the term generally refers to age-related macular degeneration (AMD or ARMD). Age-related macular degeneration begins with characteristic yellow deposits in the macula (central area of the retina which provides detailed central vision, called fovea) called drusen between the retinal pigment epithelium and the underlying choroid. Most people with these early changes (referred to as age-related maculopathy) have good vision. People with drusen can go on to develop advanced AMD. The risk is considerably higher when the drusen are large and numerous and associated with disturbance in the pigmented cell layer under the macula. Recent research suggests that large and soft drusen are related to elevated cholesterol deposits and may respond to cholesterol lowering agents or the Rheo Procedure. Advanced AMD, which is responsible for profound vision loss, has two forms: dry and wet. Central geographic atrophy, the dry form of advanced AMD, results from atrophy to the retinal pigment epithelial layer below the retina, which causes vision loss through loss of photoreceptors (rods and cones) in the central part of the eye. -

Causes of Color Blindness: Function and Failure of the Genes That Detect Color
A REVIEW ON COLOR BLINDNESS 1 Causes of Color Blindness: Function and Failure of the Genes that Detect Color Dylan Taylor A Senior Thesis submitted in partial fulfillment of the requirements for graduation in the Honors Program Liberty University Fall 2020 A REVIEW ON COLOR BLINDNESS 2 Acceptance of Senior Honors Thesis This Senior Honors Thesis is accepted in partial fulfillment of the requirements for graduation from the Honors Program of Liberty University. ______________________________ Gary D. Isaacs Jr., Ph.D. Thesis Chair ______________________________ Michael S. Price, Ph.D. Committee Member _____________________________ James H. Nutter, D.A. Honors Director ______________________________ Date A REVIEW ON COLOR BLINDNESS 3 Abstract Color blindness affects nearly 10% of the entire population, with multiple types of color blindness from various genetic mutations. In the following sections, the nature of light and how the human eye perceives light will be discussed. Afterward, the major forms of color blindness and their genetic causes will be considered. Once these genetic causes have been established, the current method for diagnosing color blindness will be investigated, followed by a discussion of the current treatments available to those with color blindness. Finally, a brief discussion will address possible future work for color blindness with the hope of finding better treatments and a future prevention. A REVIEW ON COLOR BLINDNESS 4 Causes of Color Blindness: Function and Failure of the Genes that Detect Color Introduction Without the human eye’s ability to detect color, the world would appear as dull as a black and white movie. However, despite our ability to detect color, not all humans perceive the same colors as one another. -

The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease
International Journal of Molecular Sciences Review The Alter Retina: Alternative Splicing of Retinal Genes in Health and Disease Izarbe Aísa-Marín 1,2 , Rocío García-Arroyo 1,3 , Serena Mirra 1,2 and Gemma Marfany 1,2,3,* 1 Departament of Genetics, Microbiology and Statistics, Avda. Diagonal 643, Universitat de Barcelona, 08028 Barcelona, Spain; [email protected] (I.A.-M.); [email protected] (R.G.-A.); [email protected] (S.M.) 2 Centro de Investigación Biomédica en Red Enfermedades Raras (CIBERER), Instituto de Salud Carlos III (ISCIII), Universitat de Barcelona, 08028 Barcelona, Spain 3 Institute of Biomedicine (IBUB, IBUB-IRSJD), Universitat de Barcelona, 08028 Barcelona, Spain * Correspondence: [email protected] Abstract: Alternative splicing of mRNA is an essential mechanism to regulate and increase the diversity of the transcriptome and proteome. Alternative splicing frequently occurs in a tissue- or time-specific manner, contributing to differential gene expression between cell types during development. Neural tissues present extremely complex splicing programs and display the highest number of alternative splicing events. As an extension of the central nervous system, the retina constitutes an excellent system to illustrate the high diversity of neural transcripts. The retina expresses retinal specific splicing factors and produces a large number of alternative transcripts, including exclusive tissue-specific exons, which require an exquisite regulation. In fact, a current challenge in the genetic diagnosis of inherited retinal diseases stems from the lack of information regarding alternative splicing of retinal genes, as a considerable percentage of mutations alter splicing Citation: Aísa-Marín, I.; or the relative production of alternative transcripts. Modulation of alternative splicing in the retina García-Arroyo, R.; Mirra, S.; Marfany, is also instrumental in the design of novel therapeutic approaches for retinal dystrophies, since it G. -

Addressing the Needs of Students with Color Vision Deficiencies in the Elementary School Library
Old Dominion University ODU Digital Commons Teaching & Learning Theses & Dissertations Teaching & Learning Summer 2013 Addressing the Needs of Students With Color Vision Deficiencies in the Elementary School Library Karla Bame Collins Old Dominion University Follow this and additional works at: https://digitalcommons.odu.edu/teachinglearning_etds Part of the Educational Assessment, Evaluation, and Research Commons, Elementary Education Commons, and the Library and Information Science Commons Recommended Citation Collins, Karla B.. "Addressing the Needs of Students With Color Vision Deficiencies in the Elementary School Library" (2013). Doctor of Philosophy (PhD), Dissertation, Teaching & Learning, Old Dominion University, DOI: 10.25777/ye1d-ps55 https://digitalcommons.odu.edu/teachinglearning_etds/46 This Dissertation is brought to you for free and open access by the Teaching & Learning at ODU Digital Commons. It has been accepted for inclusion in Teaching & Learning Theses & Dissertations by an authorized administrator of ODU Digital Commons. For more information, please contact [email protected]. ADDRESSING THE NEEDS OF STUDENTS WITH COLOR VISION DEFICIENCIES IN THE ELEMENTARY SCHOOL LIBRARY by Karla Bame Collins B.S. May 1991, James Madison University M.A.Ed. May 2003, College of William and Mary A Dissertation Submitted to the Faculty of Old Dominion University in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY EDUCATION OLD DOMINION UNIVERSITY August 2013 Approved by: Carol A. Doll (Committee Chair) -

BCM&School-EN-Final 3
Board of Directors BEST PRACTICES FOR SCHOOL INTEGRATION OF President STUDENTS WITH BLUE CONE MONOCHROMACY Renata Sarno, Ph.D. (BCM) Secretary Kay McCrary, Ed.D. By BCM Families Foundation - October 2015 Treasurer Barbara Sergent, MBA This document contains notes and advice collected during many years Scientific Advisory Board of firsthand experience by men who have BCM and by families with children who have BCM, most of whom live in USA and in Europe. It Jeremy Nathans, MD, Ph.D. has been written for teachers of Primary and Secondary/Elementary, The Johns Hopkins School of Middle and High Schools. We hope this document will help students Medicine, Baltimore, Maryland with BCM by giving their teachers useful information about the visual Samuel G. Jacobson, MD, Ph.D. impairments caused by BCM, plus by recommending proven strategies Scheie Eye Institute, University of to better integrate the BCM student. Philadelphia William W. Hauswirth, Ph.D. University of Florida Thanks to all the people contributing to editing this document: Kay John G. Flannery, Ph.D. McCrary Ed.D., Renata Sarno, Ph.D., Trudi Dawson, Valentina Della Volpe University of California, Berkeley Ph.D, and to all the families who sent us advice and comments. We thank Dr. Laura K. Windsor of the Low Vision Center of Indiana, because her Alessandro Iannaccone, MD,M.S. Hamilton Eye Institute,University Teacher’s Guide for Achromatopsia inspired us to write this guide for of Tennessee BCM. Bernd Wissinger, Ph.D. University of Tuebingen, Germany Communication or comments can be sent to: [email protected] – new suggestions are welcomed! Toward the cure of Blue Cone Monochromacy BCM Families Foundation PO Box 7711 Jupiter, FL 33458-7711 USA [email protected] BCM Families Foundation is recognized as a non-profit organization by the Internal Revenue www.BCMFamilies.org Service. -

Cone Targeted Gene Therapy: Animal Models
Cone Targeted Gene Therapy: Animal Models Rialto Beach, WA 2007 Which cone disease? Targets only cones Genetics are well established Animal model mimics human disease Clear clinical assay for therapy Cone genes causing Phototransduction foveal function loss 3 genes causing Achromatopsia CNGA3 CNGB3 GNAT2 J Med Genet 2002;39:656–660 Cone genes causing Phototransduction foveal function loss 3 genes causing Achromatopsia CNGA3 CNGB3 GNAT2 X-linked color blindness J Med Genet 2002;39:656–660 Cone genes causing Phototransduction foveal function loss 3 genes causing Achromatopsia CNGA3 CNGB3 GNAT2 X-linked color blindness Blue Cone Monochromacy J Med Genet 2002;39:656–660 Cone genes causing Phototransduction foveal function loss 3 genes causing Achromatopsia CNGA3 CNGB3 GNAT2 X-linked color blindness Blue Cone Monochromacy J Med Genet 2002;39:656–660 Mouse model for GNAT2 Achromatopsia Alexander, J, Umino, Y , Chang, B , Min, S , Li, Q, Timmers, A , Hawes, N, Everhart, D, Barlow, R, Pang, P and Hauswirth, WW “Restoration of Cone Function in a Mouse Model of Achromatopsia” Nature Medicine. 13:685-687 (2007). AAV5-PR2.1-mGnat2 Treatment effects Is useful vision restored? Cone ERG ~100% of normal Cone structure ~normal Mouse model for GNAT2 Achromatopsia Alexander, J, Umino, Y , Chang, B , Min, S , Li, Q, Timmers, A , Hawes, N, Everhart, D, Barlow, R, Pang, P and Hauswirth, WW “Restoration of Cone Function in a Mouse Model of Achromatopsia” Nature Medicine. 13:685-687 (2007). AAV5-PR2.1-mGnat2 Treatment effects Cone ERG ~100% of normal Cone structure ~normal Visual Acuity 0.4 0.3 0.2 Visual Acuity Visual 0.1 (cycles / degree) / (cycles 0.0 Untreated Treated Wild Type Cpfl3 Eyes Cpfl3 Eyes Eyes Nature Medicine (2007). -

Color Vision Deficiency
Color vision deficiency Description Color vision deficiency (sometimes called color blindness) represents a group of conditions that affect the perception of color. Red-green color vision defects are the most common form of color vision deficiency. Affected individuals have trouble distinguishing between some shades of red, yellow, and green. Blue-yellow color vision defects (also called tritan defects), which are rarer, cause problems with differentiating shades of blue and green and cause difficulty distinguishing dark blue from black. These two forms of color vision deficiency disrupt color perception but do not affect the sharpness of vision (visual acuity). A less common and more severe form of color vision deficiency called blue cone monochromacy causes very poor visual acuity and severely reduced color vision. Affected individuals have additional vision problems, which can include increased sensitivity to light (photophobia), involuntary back-and-forth eye movements (nystagmus) , and nearsightedness (myopia). Blue cone monochromacy is sometimes considered to be a form of achromatopsia, a disorder characterized by a partial or total lack of color vision with other vision problems. Frequency Red-green color vision defects are the most common form of color vision deficiency. This condition affects males much more often than females. Among populations with Northern European ancestry, it occurs in about 1 in 12 males and 1 in 200 females. Red- green color vision defects have a lower incidence in almost all other populations studied. Blue-yellow color vision defects affect males and females equally. This condition occurs in fewer than 1 in 10,000 people worldwide. Blue cone monochromacy is rarer than the other forms of color vision deficiency, affecting about 1 in 100,000 people worldwide.