Convergence and Divergence in the Evolution of Aquatic Birds Marcel Van Tuinen1{, Dave Brian Butvill2,Johna.W.Kirsch2 and S
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Red-Footed Booby Helper at Great Frigatebird Nests
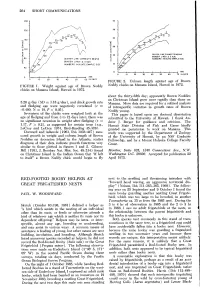
264 SHORT COMMUNICATIONS NECTS MEANS ECTS MEANS ICATE SAMPLE SIZE S.D. SAMPLE SIZE 70 IN DAYS FIGURE 2. Culmen length against age of Brown FIGURE 1. Weight against age of Brown Noddy Noddy chicks on Manana Island, Hawaii in 1972. chicks on Manana Island, Hawaii in 1972. about the thirty-fifth day; apparently Brown Noddies on Christmas Island grow more rapidly than those on 5.26 g/day (SD = 1.18 g/day), and chick growth rate Manana. More data are required for a refined analysis and fledging age were negatively correlated (r = of intraspecific variation in growth rates of Brown -0.490, N = 19, P < 0.05). Noddy young. Seventeen of the chicks were weighed both at the This paper is based upon my doctoral dissertation age of fledging and from 3 to 12 days later; there was submitted to the University of Hawaii. I thank An- no significant recession in weight after fledging (t = drew J. Berger for guidance and criticism. The 1.17, P > 0.2), as suggested for certain terns (e.g., Hawaii State Division of Fish and Game kindly LeCroy and LeCroy 1974, Bird-Banding 45:326). granted me permission to work on Manana. This Dorward and Ashmole (1963, Ibis 103b: 447) mea- study was supported by the Department of Zoology sured growth in weight and culmen length of Brown of the University of Hawaii, by an NSF Graduate Noddies on Ascension Island in the Atlantic; scatter Fellowship, and by a Mount Holyoke College Faculty diagrams of their data indicate growth functions very Grant. -

How Seabirds Plunge-Dive Without Injuries
How seabirds plunge-dive without injuries Brian Changa,1, Matthew Crosona,1, Lorian Strakerb,c,1, Sean Garta, Carla Doveb, John Gerwind, and Sunghwan Junga,2 aDepartment of Biomedical Engineering and Mechanics, Virginia Polytechnic Institute and State University, Blacksburg, VA 24061; bNational Museum of Natural History, Smithsonian Institution, Washington, DC 20560; cSetor de Ornitologia, Museu Nacional, Universidade Federal do Rio de Janeiro, São Cristóvão, Rio de Janeiro RJ 20940-040, Brazil; and dNorth Carolina Museum of Natural Sciences, Raleigh, NC 27601 Edited by David A. Weitz, Harvard University, Cambridge, MA, and approved August 30, 2016 (received for review May 27, 2016) In nature, several seabirds (e.g., gannets and boobies) dive into wa- From a mechanics standpoint, an axial force acting on a slender ter at up to 24 m/s as a hunting mechanism; furthermore, gannets body may lead to mechanical failure on the body, otherwise known and boobies have a slender neck, which is potentially the weakest as buckling. Therefore, under compressive loads, the neck is po- part of the body under compression during high-speed impact. In tentially the weakest part of the northern gannet due to its long this work, we investigate the stability of the bird’s neck during and slender geometry. Still, northern gannets impact the water at plunge-diving by understanding the interaction between the fluid up to 24 m/s without injuries (18) (see SI Appendix, Table S1 for forces acting on the head and the flexibility of the neck. First, we estimated speeds). The only reported injuries from plunge-diving use a salvaged bird to identify plunge-diving phases. -

Onetouch 4.0 Scanned Documents
/ Chapter 2 THE FOSSIL RECORD OF BIRDS Storrs L. Olson Department of Vertebrate Zoology National Museum of Natural History Smithsonian Institution Washington, DC. I. Introduction 80 II. Archaeopteryx 85 III. Early Cretaceous Birds 87 IV. Hesperornithiformes 89 V. Ichthyornithiformes 91 VI. Other Mesozojc Birds 92 VII. Paleognathous Birds 96 A. The Problem of the Origins of Paleognathous Birds 96 B. The Fossil Record of Paleognathous Birds 104 VIII. The "Basal" Land Bird Assemblage 107 A. Opisthocomidae 109 B. Musophagidae 109 C. Cuculidae HO D. Falconidae HI E. Sagittariidae 112 F. Accipitridae 112 G. Pandionidae 114 H. Galliformes 114 1. Family Incertae Sedis Turnicidae 119 J. Columbiformes 119 K. Psittaciforines 120 L. Family Incertae Sedis Zygodactylidae 121 IX. The "Higher" Land Bird Assemblage 122 A. Coliiformes 124 B. Coraciiformes (Including Trogonidae and Galbulae) 124 C. Strigiformes 129 D. Caprimulgiformes 132 E. Apodiformes 134 F. Family Incertae Sedis Trochilidae 135 G. Order Incertae Sedis Bucerotiformes (Including Upupae) 136 H. Piciformes 138 I. Passeriformes 139 X. The Water Bird Assemblage 141 A. Gruiformes 142 B. Family Incertae Sedis Ardeidae 165 79 Avian Biology, Vol. Vlll ISBN 0-12-249408-3 80 STORES L. OLSON C. Family Incertae Sedis Podicipedidae 168 D. Charadriiformes 169 E. Anseriformes 186 F. Ciconiiformes 188 G. Pelecaniformes 192 H. Procellariiformes 208 I. Gaviiformes 212 J. Sphenisciformes 217 XI. Conclusion 217 References 218 I. Introduction Avian paleontology has long been a poor stepsister to its mammalian counterpart, a fact that may be attributed in some measure to an insufRcien- cy of qualified workers and to the absence in birds of heterodont teeth, on which the greater proportion of the fossil record of mammals is founded. -

MAGNIFICENT FRIGATEBIRD Fregata Magnificens
PALM BEACH DOLPHIN PROJECT FACT SHEET The Taras Oceanographic Foundation 5905 Stonewood Court - Jupiter, FL 33458 - (561-762-6473) [email protected] MAGNIFICENT FRIGATEBIRD Fregata magnificens CLASS: Aves ORDER: Suliformes FAMILY: Fregatidae GENUS: Fregata SPECIES: magnificens A long-winged, fork-tailed bird of tropical oceans, the Magnificent Frigatebird is an agile flier that snatches food off the surface of the ocean and steals food from other birds. It breeds mostly south of the United States, but wanders northward along the coasts during nonbreeding season. Physical Appearance: Frigate birds are the only seabirds where the male and female look strikingly different. All have pre- dominantly black plumage, long, deeply forked tails and long hooked bills. Females have white underbellies and males have a distinctive red throat pouch, which they inflate during the breeding season to attract females Their wings are long and pointed and can span up to 2.3 meters (7.5 ft), the largest wing area to body weight ratio of any bird. These birds are about 35-45 inches ((89 to 114 cm) in length, and weight between 35 and 67 oz (1000-1900 g). The bones of frigate birds are markedly pneumatic (filled with air), making them very light and contribute only 5% to total body weight. The pectoral girdle (shoulder joint) is strong as its bones are fused. Habitat: Frigate birds are found across all tropical oceans. Breeding habitats include mangrove cays on coral reefs, and decidu- ous trees and bushes on dry islands. Feeding range while breeding includes shallow water within lagoons, coral reefs, and deep ocean out of sight of land. -

Bontebok Birds
Birds recorded in the Bontebok National Park 8 Little Grebe 446 European Roller 55 White-breasted Cormorant 451 African Hoopoe 58 Reed Cormorant 465 Acacia Pied Barbet 60 African Darter 469 Red-fronted Tinkerbird * 62 Grey Heron 474 Greater Honeyguide 63 Black-headed Heron 476 Lesser Honeyguide 65 Purple Heron 480 Ground Woodpecker 66 Great Egret 486 Cardinal Woodpecker 68 Yellow-billed Egret 488 Olive Woodpecker 71 Cattle Egret 494 Rufous-naped Lark * 81 Hamerkop 495 Cape Clapper Lark 83 White Stork n/a Agulhas Longbilled Lark 84 Black Stork 502 Karoo Lark 91 African Sacred Ibis 504 Red Lark * 94 Hadeda Ibis 506 Spike-heeled Lark 95 African Spoonbill 507 Red-capped Lark 102 Egyptian Goose 512 Thick-billed Lark 103 South African Shelduck 518 Barn Swallow 104 Yellow-billed Duck 520 White-throated Swallow 105 African Black Duck 523 Pearl-breasted Swallow 106 Cape Teal 526 Greater Striped Swallow 108 Red-billed Teal 529 Rock Martin 112 Cape Shoveler 530 Common House-Martin 113 Southern Pochard 533 Brown-throated Martin 116 Spur-winged Goose 534 Banded Martin 118 Secretarybird 536 Black Sawwing 122 Cape Vulture 541 Fork-tailed Drongo 126 Black (Yellow-billed) Kite 547 Cape Crow 127 Black-shouldered Kite 548 Pied Crow 131 Verreauxs' Eagle 550 White-necked Raven 136 Booted Eagle 551 Grey Tit 140 Martial Eagle 557 Cape Penduline-Tit 148 African Fish-Eagle 566 Cape Bulbul 149 Steppe Buzzard 572 Sombre Greenbul 152 Jackal Buzzard 577 Olive Thrush 155 Rufous-chested Sparrowhawk 582 Sentinel Rock-Thrush 158 Black Sparrowhawk 587 Capped Wheatear -

Phylogenetic Patterns of Size and Shape of the Nasal Gland Depression in Phalacrocoracidae
PHYLOGENETIC PATTERNS OF SIZE AND SHAPE OF THE NASAL GLAND DEPRESSION IN PHALACROCORACIDAE DOUGLAS SIEGEL-CAUSEY Museumof NaturalHistory and Department of Systematicsand Ecology, University of Kansas, Lawrence, Kansas 66045-2454 USA ABSTRACT.--Nasalglands in Pelecaniformesare situatedwithin the orbit in closelyfitting depressions.Generally, the depressionsare bilobedand small,but in Phalacrocoracidaethey are more diversein shapeand size. Cormorants(Phalacrocoracinae) have small depressions typical of the order; shags(Leucocarboninae) have large, single-lobeddepressions that extend almost the entire length of the frontal. In all PhalacrocoracidaeI examined, shape of the nasalgland depressiondid not vary betweenfreshwater and marine populations.A general linear model detectedstrongly significant effectsof speciesidentity and gender on size of the gland depression.The effectof habitat on size was complexand was detectedonly as a higher-ordereffect. Age had no effecton size or shapeof the nasalgland depression.I believe that habitat and diet are proximateeffects. The ultimate factorthat determinessize and shape of the nasalgland within Phalacrocoracidaeis phylogenetichistory. Received 28 February1989, accepted1 August1989. THE FIRSTinvestigations of the nasal glands mon (e.g.Technau 1936, Zaks and Sokolova1961, of water birds indicated that theseglands were Thomson and Morley 1966), and only a few more developed in species living in marine studies have focused on the cranial structure habitats than in species living in freshwater associatedwith the nasal gland (Marpies 1932; habitats (Heinroth and Heinroth 1927, Marpies Bock 1958, 1963; Staaland 1967; Watson and Di- 1932). Schildmacher (1932), Technau (1936), and voky 1971; Lavery 1972). othersshowed that the degree of development Unlike most other birds, Pelecaniformes have among specieswas associatedwith habitat. Lat- nasal glands situated in depressionsfound in er experimental studies (reviewed by Holmes the anteromedialroof of the orbit (Siegel-Cau- and Phillips 1985) established the role of the sey 1988). -

A Skull of a Very Large Crane from the Late Miocene of Southern Germany, with Notes on the Phylogenetic Interrelationships of Extant Gruinae
Journal of Ornithology https://doi.org/10.1007/s10336-020-01799-0 ORIGINAL ARTICLE A skull of a very large crane from the late Miocene of Southern Germany, with notes on the phylogenetic interrelationships of extant Gruinae Gerald Mayr1 · Thomas Lechner2 · Madelaine Böhme2 Received: 3 June 2020 / Revised: 1 July 2020 / Accepted: 2 July 2020 © The Author(s) 2020 Abstract We describe a partial skull of a very large crane from the early late Miocene (Tortonian) hominid locality Hammerschmiede in southern Germany, which is the oldest fossil record of the Gruinae (true cranes). The fossil exhibits an unusual preservation in that only the dorsal portions of the neurocranium and beak are preserved. Even though it is, therefore, very fragmentary, two morphological characteristics are striking and of paleobiological signifcance: its large size and the very long beak. The fossil is from a species the size of the largest extant cranes and represents the earliest record of a large-sized crane in Europe. Overall, the specimen resembles the skull of the extant, very long-beaked Siberian Crane, Leucogeranus leucogeranus, but its afnities within Gruinae cannot be determined owing to the incomplete preservation. Judging from its size, the fossil may possibly belong to the very large “Grus” pentelici, which stems from temporally and geographically proximate sites. The long beak of the Hammerschmiede crane conforms to an open freshwater paleohabitat, which prevailed at the locality. Keywords Aves · Evolution · Gruidae · Systematics · Tortonian Zusammenfassung Ein Schädel eines sehr großen Kranichs aus dem Obermiozän Süddeutschlands, mit Bemerkungen zu den Verwandtschaftsbeziehungen heutiger Gruinae Wir beschreiben einen partiell erhaltenen Schädel eines sehr großen Kranichs aus dem frühen Obermiozän (Tortonium) der Hominiden-Fundstelle Hammerschmiede in Süddeutschland, welcher den ältesten Fossilnachweis der Gruinae (echte Kraniche) repräsentiert. -

P0529-P0540.Pdf
RESPONSES TO HIGH TEMPERATURE IN NESTLING DOUBLE-CRESTED AND PELAGIC CORMORANTS ROBERT C. L^$IEWSKI AND GREGORY K. SNYDER ADULTand nestlingcormorants are often subjectto overheatingfrom insolationat the nest. Their generallydark plumage,exposed nest sites, and reradiation from surroundingrocks aggravate the thermal stress. Young nestlingsmust be shieldedfrom the sun by their parents. Older nestlingsand adults compensatefor heat gain throughbehavioral adjust- mentsand modulationof evaporativecooling by pantingand gular flutter- ing. This study was undertakento examinesome of the responsesto high temperaturein nestlingsof two speciesof cormorants,the Double-crested Cormorant,Phalacrocorax auritus, and the PelagicCormorant, P. pelagicus. The evaporative cooling responsesin birds have been studied in some detail in recent years (see Bartholomewet al., 1962; Lasiewski et al., 1966; Bartholomewet al., 1968; Calder and Schmidt-Nielsen,1968, for more detailed discussions),although much still remains to be learned. MATERIALS AND •VIETI-IODS The nestling cormorantsused in this study (four Phalacrocoraxpelagicus and four P. auritus) were captured from nests on rocky islands off the northwest coast of Washington. As their dates of hatchingwere not known it was impossibleto provide exact ages. From comparisonsof feather developmentwith descriptionsin the literature (Bent, 1922; Palmer, 1962), we judged that the pe'lagicuschicks were approximately 5, 5, 6, and 6 weeksold, while'the auritus chickswere 3.5, 3.5, 4.5, and 6 weeksof age upon capture. The nestlingswere taken to the laboratoriesat Friday Harbor, Washington on the day of capture and housed in three 4' X 4' X 4' chicken wire cages. The cageswere equippedwith plywood platforms for the birds to sit on and coveredon top and two sides to shield birds from wind and rain. -

The Hamerkop Colorado Tropics 2
November/December 2009 Volume 6 Issue 2 a i k e n a u d u b o n s o c i e t y Bird Profiles: I n s I d e t h I s I s s u e Coming Program 2 The Hamerkop Colorado Tropics 2 n case you’re wondering, this is a Hamerkop (also known Field Trips & Events 3 zmann Ias a Hammerhead, Anvil Bird, or Umber Bird, among L other aliases). I found him at the Denver Zoo, where he was Lesser Prairie-Chickens 4 e ho making quite the racket. Apparently this is somewhat unusual li as they are typically pretty quiet. Don’t Feed the Bears 4 These are really strange birds. According to Wikipedia, my photo: Les Are You a Beak Geek? 5 favorite source of potentially correct information, “One unusual feature is that up to ten birds join in ‘ceremonies’ in Subscription Form 5 which they run circles around each other, all calling loudly, raising their crests, [and] fluttering their wings.” Reminds Wesley the Owl back me of my high school football team. If that isn’t weird enough, consider their nesting habits. It would seem that Hamerkops are really into nest building. Using c o m I ng programs perhaps 10,000 sticks, they construct a huge edifice more than four feet across. The sticks, cemented together with mud, are November 19 formed into walls and covered with a domed roof. This bird “Birds of Bolivia” mansion is accessed by a tunnel, also mortared with mud, presented by John Drummond and decorated with the latest in colorful objets d’ art. -

Birds (DNA'dna Hybridization/Mtdna Sequences/Phylogeny/Systematics)
Proc. Natl. Acad. Sci. USA Vol. 91, pp. 9861-9865, October 1994 Evolution Molecules vs. morphology in avian evolution: The case of the "pelecaniform" birds (DNA'DNA hybridization/mtDNA sequences/phylogeny/systematics) S. BLAIR HEDGES* AND CHARLES G. SIBLEyt *Department of Biology and Institute of Molecular Evolutionary Genetics, 208 Mueller Laboratory, Pennsylvania State University, University Park, PA 16802; and t433 Woodley Place, Santa Rosa, CA 95409 Contributed by Charles G. Sibley, June 20, 1994 ABSTRACT The traditional avian Order Pelecaniformes the three front toes has evolved in groups with separate is composed of birds with all four toes connected by a web. This origins-e.g., ducks, gulls, flamingos, and albatrosses. Could "totipalmate" condition is found in ca. 66 living species: 8 the totipalmate condition, which occurs in fewer species, also pelicans (Pelecanus), 9 boobies and gannets (Sula, Papasula, have multiple origins? Sibley and Ahlquist (2) reviewed the Morus), ca. 37 cormorants (Phalacrocorax) , 4 anhingas or literature from 1758 to 1990. darters (Anhinga), 5 frigatebirds (Fregata), and 3 tropicbirds There have been many morphological studies of the pele (Phaethon). Several additional characters are shared by these caniforms; those of Lanham (3), Saiff(4), and Cracraft (5) are genera, and their monophyly has been assumed since the among the most recent. Lanham (3) recognized their diversity beginning of modern zoological nomenclature. Most ornithol but concluded that the totipalmate birds form a natural order. ogists classify these genera as an order, although tropicbirds He assigned Phaethon and Fregata to separate suborders, have been viewed as related to terns, and frigatebirds as the other genera to the suborder Pelecani, and suggested that relatives of the petrels and albatrosses. -

Family Friendly Activities All Our Activities Can Be Adapted for Children and Young People from Age 5 up to 18. It's Best To
Family Friendly Activities All our activities can be adapted for children and young people from age 5 up to 18. It’s best to book ahead so we can make sure we can cater for your group and learn more about your interests. Family groups or adults are also welcome to arrange special activity sessions too if you would like to explore more! Each activity session costs 10,000 Rwf in addition to the Umusambi Village entrance fee. Looking After Cranes Have you ever wondered how the cranes are looked after at Umusambi Village? Join one of our experienced wildlife veterinarians for a behind the scenes tour of the sanctuary. You will find out about the work of the vets and the equipment they use to examine and treat cranes, as well as getting up close to some of the cranes. You will learn about the different body parts and how cranes differ from people as well as what they need to be healthy. We will use binoculars to check on the cranes at Umusambi Village and learn how some of them came to be at the sanctuary. We have a wide variety of games and activities to suit different age groups. Day: Only available on Fridays. Duration: 1hour 30 Seeds, Plants and Trees Plants and trees are so important to our environment. Would you like to know more about special plants and trees in Rwanda and find out about some of the stories behind them? At Umusambi Village we have a tree nursery where we grow trees from tiny seeds to small trees. -

How Birds Combat Ectoparasites
The Open Ornithology Journal, 2010, 3, 41-71 41 Open Access How Birds Combat Ectoparasites Dale H. Clayton*,1, Jennifer A. H. Koop1, Christopher W. Harbison1,2, Brett R. Moyer1,3 and Sarah E. Bush1,4 1Department of Biology, University of Utah, Salt Lake City, UT 84112, USA; 2Current address: Biology Department, Siena College, Loudonville, NY, 12211, USA; 3Current address: Providence Day School, Charlotte, NC, 28270, USA; 4Natural History Museum and Biodiversity Research Center, University of Kansas, Lawrence, Kansas 66045, USA Abstract: Birds are plagued by an impressive diversity of ectoparasites, ranging from feather-feeding lice, to feather- degrading bacteria. Many of these ectoparasites have severe negative effects on host fitness. It is therefore not surprising that selection on birds has favored a variety of possible adaptations for dealing with ectoparasites. The functional signifi- cance of some of these defenses has been well documented. Others have barely been studied, much less tested rigorously. In this article we review the evidence - or lack thereof - for many of the purported mechanisms birds have for dealing with ectoparasites. We concentrate on features of the plumage and its components, as well as anti-parasite behaviors. In some cases, we present original data from our own recent work. We make recommendations for future studies that could im- prove our understanding of this poorly known aspect of avian biology. Keywords: Grooming, preening, dusting, sunning, molt, oil, anting, fumigation. INTRODUCTION 2) Mites and ticks (Acari): many families [6-9]. As a class, birds (Aves) are the most thoroughly studied 3) Leeches: four families [10]. group of organisms on earth.