UC Santa Barbara Dissertation Template
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Qt9z7703dj.Pdf
UC San Diego UC San Diego Previously Published Works Title Phylogeny and biogeography of a shallow water fish clade (Teleostei: Blenniiformes) Permalink https://escholarship.org/uc/item/9z7703dj Journal BMC Evolutionary Biology, 13(1) ISSN 1471-2148 Authors Lin, Hsiu-Chin Hastings, Philip A Publication Date 2013-09-25 DOI http://dx.doi.org/10.1186/1471-2148-13-210 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Lin and Hastings BMC Evolutionary Biology 2013, 13:210 http://www.biomedcentral.com/1471-2148/13/210 RESEARCH ARTICLE Open Access Phylogeny and biogeography of a shallow water fish clade (Teleostei: Blenniiformes) Hsiu-Chin Lin1,2* and Philip A Hastings1 Abstract Background: The Blenniiformes comprises six families, 151 genera and nearly 900 species of small teleost fishes closely associated with coastal benthic habitats. They provide an unparalleled opportunity for studying marine biogeography because they include the globally distributed families Tripterygiidae (triplefin blennies) and Blenniidae (combtooth blennies), the temperate Clinidae (kelp blennies), and three largely Neotropical families (Labrisomidae, Chaenopsidae, and Dactyloscopidae). However, interpretation of these distributional patterns has been hindered by largely unresolved inter-familial relationships and the lack of evidence of monophyly of the Labrisomidae. Results: We explored the phylogenetic relationships of the Blenniiformes based on one mitochondrial (COI) and four nuclear (TMO-4C4, RAG1, Rhodopsin, and Histone H3) loci for 150 blenniiform species, and representative outgroups (Gobiesocidae, Opistognathidae and Grammatidae). According to the consensus of Bayesian Inference, Maximum Likelihood, and Maximum Parsimony analyses, the monophyly of the Blenniiformes and the Tripterygiidae, Blenniidae, Clinidae, and Dactyloscopidae is supported. -

CHECKLIST and BIOGEOGRAPHY of FISHES from GUADALUPE ISLAND, WESTERN MEXICO Héctor Reyes-Bonilla, Arturo Ayala-Bocos, Luis E
ReyeS-BONIllA eT Al: CheCklIST AND BIOgeOgRAphy Of fISheS fROm gUADAlUpe ISlAND CalCOfI Rep., Vol. 51, 2010 CHECKLIST AND BIOGEOGRAPHY OF FISHES FROM GUADALUPE ISLAND, WESTERN MEXICO Héctor REyES-BONILLA, Arturo AyALA-BOCOS, LUIS E. Calderon-AGUILERA SAúL GONzáLEz-Romero, ISRAEL SáNCHEz-ALCántara Centro de Investigación Científica y de Educación Superior de Ensenada AND MARIANA Walther MENDOzA Carretera Tijuana - Ensenada # 3918, zona Playitas, C.P. 22860 Universidad Autónoma de Baja California Sur Ensenada, B.C., México Departamento de Biología Marina Tel: +52 646 1750500, ext. 25257; Fax: +52 646 Apartado postal 19-B, CP 23080 [email protected] La Paz, B.C.S., México. Tel: (612) 123-8800, ext. 4160; Fax: (612) 123-8819 NADIA C. Olivares-BAñUELOS [email protected] Reserva de la Biosfera Isla Guadalupe Comisión Nacional de áreas Naturales Protegidas yULIANA R. BEDOLLA-GUzMáN AND Avenida del Puerto 375, local 30 Arturo RAMíREz-VALDEz Fraccionamiento Playas de Ensenada, C.P. 22880 Universidad Autónoma de Baja California Ensenada, B.C., México Facultad de Ciencias Marinas, Instituto de Investigaciones Oceanológicas Universidad Autónoma de Baja California, Carr. Tijuana-Ensenada km. 107, Apartado postal 453, C.P. 22890 Ensenada, B.C., México ABSTRACT recognized the biological and ecological significance of Guadalupe Island, off Baja California, México, is Guadalupe Island, and declared it a Biosphere Reserve an important fishing area which also harbors high (SEMARNAT 2005). marine biodiversity. Based on field data, literature Guadalupe Island is isolated, far away from the main- reviews, and scientific collection records, we pres- land and has limited logistic facilities to conduct scien- ent a comprehensive checklist of the local fish fauna, tific studies. -

Fish Assemblage of the Mamanguape Environmental Protection Area, NE Brazil: Abundance, Composition and Microhabitat Availability Along the Mangrove-Reef Gradient
Neotropical Ichthyology, 10(1): 109-122, 2012 Copyright © 2012 Sociedade Brasileira de Ictiologia Fish assemblage of the Mamanguape Environmental Protection Area, NE Brazil: abundance, composition and microhabitat availability along the mangrove-reef gradient Josias Henrique de Amorim Xavier1, Cesar Augusto Marcelino Mendes Cordeiro2, Gabrielle Dantas Tenório1, Aline de Farias Diniz1, Eugenio Pacelli Nunes Paulo Júnior1, Ricardo S. Rosa1 and Ierecê Lucena Rosa1 Reefs, mangroves and seagrass biotopes often occur in close association, forming a complex and highly productive ecosystem that provide significant ecologic and economic goods and services. Different anthropogenic disturbances are increasingly affecting these tropical coastal habitats leading to growing conservation concern. In this field-based study, we used a visual census technique (belt transects 50 m x 2 m) to investigate the interactions between fishes and microhabitats at the Mamanguape Mangrove-Reef system, NE Brazil. Overall, 144 belt transects were performed from October 2007 to September 2008 to assess the structure of the fish assemblage. Fish trophic groups and life stage (juveniles and adults) were recorded according to literature, the percent cover of the substrate was estimated using the point contact method. Our results revealed that fish composition gradually changed from the Estuarine to the Reef zone, and that fish assemblage was strongly related to the microhabitat availability, as suggested by the predominance of carnivores at the Estuarine zone and presence of herbivores at the Reef zone. Fish abundance and diversity were higher in the Reef zone and estuary margins, highlighting the importance of structural complexity. A pattern of nursery area utilization, with larger specimens at the Transition and Reef Zone and smaller individuals at the Estuarine zone, was recorded for Abudefduf saxatilis, Anisotremus surinamensis, Lutjanus alexandrei, and Lutjanus jocu. -

First Molecular Data and Morphological Re-Description of Two
Journal of King Saud University – Science 33 (2021) 101290 Contents lists available at ScienceDirect Journal of King Saud University – Science journal homepage: www.sciencedirect.com Original article First molecular data and morphological re-description of two copepod species, Hatschekia sargi and Hatschekia leptoscari, as parasites on Parupeneus rubescens in the Arabian Gulf ⇑ Saleh Al-Quraishy a, , Mohamed A. Dkhil a,b, Nawal Al-Hoshani a, Wejdan Alhafidh a, Rewaida Abdel-Gaber a,c a Zoology Department, College of Science, King Saud University, Riyadh, Saudi Arabia b Department of Zoology and Entomology, Faculty of Science, Helwan University, Cairo, Egypt c Zoology Department, Faculty of Science, Cairo University, Cairo, Egypt article info abstract Article history: Little information is available about the biodiversity of parasitic copepods in the Arabian Gulf. The pre- Received 6 September 2020 sent study aimed to provide new information about different parasitic copepods gathered from Revised 30 November 2020 Parupeneus rubescens caught in the Arabian Gulf (Saudi Arabia). Copepods collected from the infected fish Accepted 9 December 2020 were studied using light microscopy and scanning electron microscopy and then examined using stan- dard staining and measuring techniques. Phylogenetic analyses were conducted based on the partial 28S rRNA gene sequences from other copepod species retrieved from GenBank. Two copepod species, Keywords: Hatschekia sargi Brian, 1902 and Hatschekia leptoscari Yamaguti, 1939, were identified as naturally 28S rRNA gene infected the gills of fish. Here we present a phylogenetic analysis of the recovered copepod species to con- Arabian Gulf Hatschekiidae firm their taxonomic position in the Hatschekiidae family within Siphonostomatoida and suggest the Marine fish monophyletic origin this family. -
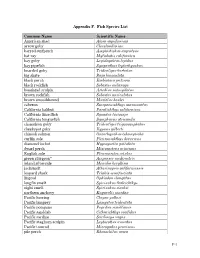
Appendix E: Fish Species List
Appendix F. Fish Species List Common Name Scientific Name American shad Alosa sapidissima arrow goby Clevelandia ios barred surfperch Amphistichus argenteus bat ray Myliobatis californica bay goby Lepidogobius lepidus bay pipefish Syngnathus leptorhynchus bearded goby Tridentiger barbatus big skate Raja binoculata black perch Embiotoca jacksoni black rockfish Sebastes melanops bonehead sculpin Artedius notospilotus brown rockfish Sebastes auriculatus brown smoothhound Mustelus henlei cabezon Scorpaenichthys marmoratus California halibut Paralichthys californicus California lizardfish Synodus lucioceps California tonguefish Symphurus atricauda chameleon goby Tridentiger trigonocephalus cheekspot goby Ilypnus gilberti chinook salmon Oncorhynchus tshawytscha curlfin sole Pleuronichthys decurrens diamond turbot Hypsopsetta guttulata dwarf perch Micrometrus minimus English sole Pleuronectes vetulus green sturgeon* Acipenser medirostris inland silverside Menidia beryllina jacksmelt Atherinopsis californiensis leopard shark Triakis semifasciata lingcod Ophiodon elongatus longfin smelt Spirinchus thaleichthys night smelt Spirinchus starksi northern anchovy Engraulis mordax Pacific herring Clupea pallasi Pacific lamprey Lampetra tridentata Pacific pompano Peprilus simillimus Pacific sanddab Citharichthys sordidus Pacific sardine Sardinops sagax Pacific staghorn sculpin Leptocottus armatus Pacific tomcod Microgadus proximus pile perch Rhacochilus vacca F-1 plainfin midshipman Porichthys notatus rainwater killifish Lucania parva river lamprey Lampetra -

Environmental DNA Reveals the Fine-Grained and Hierarchical
www.nature.com/scientificreports OPEN Environmental DNA reveals the fne‑grained and hierarchical spatial structure of kelp forest fsh communities Thomas Lamy 1,2*, Kathleen J. Pitz 3, Francisco P. Chavez3, Christie E. Yorke1 & Robert J. Miller1 Biodiversity is changing at an accelerating rate at both local and regional scales. Beta diversity, which quantifes species turnover between these two scales, is emerging as a key driver of ecosystem function that can inform spatial conservation. Yet measuring biodiversity remains a major challenge, especially in aquatic ecosystems. Decoding environmental DNA (eDNA) left behind by organisms ofers the possibility of detecting species sans direct observation, a Rosetta Stone for biodiversity. While eDNA has proven useful to illuminate diversity in aquatic ecosystems, its utility for measuring beta diversity over spatial scales small enough to be relevant to conservation purposes is poorly known. Here we tested how eDNA performs relative to underwater visual census (UVC) to evaluate beta diversity of marine communities. We paired UVC with 12S eDNA metabarcoding and used a spatially structured hierarchical sampling design to assess key spatial metrics of fsh communities on temperate rocky reefs in southern California. eDNA provided a more‑detailed picture of the main sources of spatial variation in both taxonomic richness and community turnover, which primarily arose due to strong species fltering within and among rocky reefs. As expected, eDNA detected more taxa at the regional scale (69 vs. 38) which accumulated quickly with space and plateaued at only ~ 11 samples. Conversely, the discovery rate of new taxa was slower with no sign of saturation for UVC. -

(<I>Anisotremus Virginicus,</I> Haemulidae) And
BULLETIN OF MARINE SCIENCE, 34(1): 21-59,1984 DESCRIPTION OF PORKFISH LARVAE (ANISOTREMUS VIRGINICUS, HAEMULIDAE) AND THEIR OSTEOLOGICAL DEVELOPMENT Thomas Potthoff, Sharon Kelley, Martin Moe and Forrest Young ABSTRACT Wild-caught adult porkfish (Anisotremus virginicus, Haemulidae) were spawned in the laboratory and their larvae reared. A series of 35 larvae 2.4 mm NL to 21.5 mm SL from 2 to 30 days old or older (larvae of unknown age) was studied for pigmentation characteristics. Cleared and stained specimens were examined for meristic and osteological development. Cartilaginous neural and haemal arches develop first anteriorly, at the center, and posteriorly, above and below the notochord, but ossification of the vertebral column is from anterior in a posterior direction. Epipleural rib pairs develop from bone, but pleural rib pairs develop from cartilage first and then ossify. The second dorsal, anal and caudal fins develop rays first and simultaneously, followed by first dorsal fin spine development. The pectoral and pelvic fins are the last of all fins to develop rays. All bones basic to a perciform pectoral girdle develop with cartilaginous radials present between the pectoral fin ray bases. Development and structure of pre dorsal bones and dorsal and anal fin pterygiophores were studied. All bones basic to a perciform caudal complex developed and no fusion of any of these bones was observed in the adults. Radial cartilages developed ventrad in the hypural complex. The hyoid arches originated from cartilage but the branchiostegal rays formed from bone. The development and anatomy of the branchial skeleton were studied. Spines develop on the four bones of the opercular series in larvae and juveniles but are absent in the adults. -

Diversity of Norwegian Sea Slugs (Nudibranchia): New Species to Norwegian Coastal Waters and New Data on Distribution of Rare Species
Fauna norvegica 2013 Vol. 32: 45-52. ISSN: 1502-4873 Diversity of Norwegian sea slugs (Nudibranchia): new species to Norwegian coastal waters and new data on distribution of rare species Jussi Evertsen1 and Torkild Bakken1 Evertsen J, Bakken T. 2013. Diversity of Norwegian sea slugs (Nudibranchia): new species to Norwegian coastal waters and new data on distribution of rare species. Fauna norvegica 32: 45-52. A total of 5 nudibranch species are reported from the Norwegian coast for the first time (Doridoxa ingolfiana, Goniodoris castanea, Onchidoris sparsa, Eubranchus rupium and Proctonotus mucro- niferus). In addition 10 species that can be considered rare in Norwegian waters are presented with new information (Lophodoris danielsseni, Onchidoris depressa, Palio nothus, Tritonia griegi, Tritonia lineata, Hero formosa, Janolus cristatus, Cumanotus beaumonti, Berghia norvegica and Calma glau- coides), in some cases with considerable changes to their distribution. These new results present an update to our previous extensive investigation of the nudibranch fauna of the Norwegian coast from 2005, which now totals 87 species. An increase in several new species to the Norwegian fauna and new records of rare species, some with considerable updates, in relatively few years results mainly from sampling effort and contributions by specialists on samples from poorly sampled areas. doi: 10.5324/fn.v31i0.1576. Received: 2012-12-02. Accepted: 2012-12-20. Published on paper and online: 2013-02-13. Keywords: Nudibranchia, Gastropoda, taxonomy, biogeography 1. Museum of Natural History and Archaeology, Norwegian University of Science and Technology, NO-7491 Trondheim, Norway Corresponding author: Jussi Evertsen E-mail: [email protected] IntRODUCTION the main aims. -

Distribution, Abundance, and Biomass of Giant Sea Bass (Stereolepis Gigas) Off Santa Catalina Island, California, 2014-2015
Bull. Southern California Acad. Sci. 115(1), 2016, pp. 1–14 E Southern California Academy of Sciences, 2016 The Return of the King of the Kelp Forest: Distribution, Abundance, and Biomass of Giant Sea Bass (Stereolepis gigas) off Santa Catalina Island, California, 2014-2015 Parker H. House*, Brian L.F. Clark, and Larry G. Allen California State University, Northridge, Department of Biology, 18111 Nordhoff St., Northridge, CA, 91330 Abstract.—It is rare to find evidence of top predators recovering after being negatively affected by overfishing. However, recent findings suggest a nascent return of the critically endangered giant sea bass (Stereolepis gigas) to southern California. To provide the first population assessment of giant sea bass, surveys were conducted during the 2014/2015 summers off Santa Catalina Island, CA. Eight sites were surveyed on both the windward and leeward side of Santa Catalina Island every two weeks from June through August. Of the eight sites, three aggregations were identified at Goat Harbor, The V’s, and Little Harbor, CA. These three aggregation sites, the largest containing 24 individuals, contained a mean stock biomass of 19.6 kg/1000 m2 over both summers. Over the course of both summers the giant sea bass population was primarily made up of 1.2 - 1.3 m TL individuals with several small and newly mature fish observed in aggregations. Comparison to historical data for the island suggests giant sea bass are recovering, but have not reached pre-exploitation levels. The giant sea bass (Stereolepis gigas) is the largest teleost to inhabit nearshore rocky reefs and kelp forests in the northeastern Pacific (Hawk and Allen 2014). -

Catching the Complexity of Salmon-Louse Interactions
Fish and Shellfish Immunology 90 (2019) 199–209 Contents lists available at ScienceDirect Fish and Shellfish Immunology journal homepage: www.elsevier.com/locate/fsi Full length article Catching the complexity of salmon-louse interactions T ∗ Cristian Gallardo-Escáratea, , Valentina Valenzuela-Muñoza, Gustavo Núñez-Acuñaa, Crisleri Carreraa, Ana Teresa Gonçalvesa, Diego Valenzuela-Mirandaa, Bárbara P. Benaventea, Steven Robertsb a Interdisciplinary Center for Aquaculture Research, Laboratory of Biotechnology and Aquatic Genomics, Department of Oceanography, Universidad de Concepción, Concepción, Chile b School of Aquatic and Fishery Sciences (SAFS), University of Washington, Seattle, USA ABSTRACT The study of host-parasite relationships is an integral part of the immunology of aquatic species, where the complexity of both organisms has to be overlayed with the lifecycle stages of the parasite and immunological status of the host. A deep understanding of how the parasite survives in its host and how they display molecular mechanisms to face the immune system can be applied for novel parasite control strategies. This review highlights current knowledge about salmon and sea louse, two key aquatic animals for aquaculture research worldwide. With the aim to catch the complexity of the salmon-louse interactions, molecular information gleaned through genomic studies are presented. The host recognition system and the chemosensory receptors found in sea lice reveal complex molecular components, that in turn, can be disrupted through specific molecules such as non-coding RNAs. 1. Introduction worldwide the most studied sea lice species are Lepeophtheirus salmonis and Caligus rogercresseyi [8–10]. Annually the salmon industry presents Host-parasite interactions are a complex relationship where one estimated losses of US $ 480 million, representing between 4 and 10% organism benefits the other and often where there is a negative impact of production costs [10]. -

Terrestrial and Marine Biological Resource Information
APPENDIX C Terrestrial and Marine Biological Resource Information Appendix C1 Resource Agency Coordination Appendix C2 Marine Biological Resources Report APPENDIX C1 RESOURCE AGENCY COORDINATION 1 The ICF terrestrial biological team coordinated with relevant resource agencies to discuss 2 sensitive biological resources expected within the terrestrial biological study area (BSA). 3 A summary of agency communications and site visits is provided below. 4 California Department of Fish and Wildlife: On July 30, 2020, ICF held a conference 5 call with Greg O’Connell (Environmental Scientist) and Corianna Flannery (Environmental 6 Scientist) to discuss Project design and potential biological concerns regarding the 7 Eureka Subsea Fiber Optic Cables Project (Project). Mr. O’Connell discussed the 8 importance of considering the western bumble bee. Ms. Flannery discussed the 9 importance of the hard ocean floor substrate and asked how the cable would be secured 10 to the ocean floor to reduce or eliminate scour. The western bumble bee has been 11 evaluated in the Biological Resources section of the main document, and direct and 12 indirect impacts are avoided. The Project Description describes in detail how the cable 13 would be installed on the ocean floor, the importance of the hard bottom substrate, and 14 the need for avoidance. 15 Consultation Outcomes: 16 • The Project was designed to avoid hard bottom substrate, and RTI Infrastructure 17 (RTI) conducted surveys of the ocean floor to ensure that proper routing of the 18 cable would occur. 19 • Ms. Flannery will be copied on all communications with the National Marine 20 Fisheries Service 21 California Department of Fish and Wildlife: On August 7, 2020, ICF held a conference 22 call with Greg O’Connell to discuss a site assessment and survey approach for the 23 western bumble bee. -

Vii Fishery-At-A-Glance: California Sheephead
Fishery-at-a-Glance: California Sheephead (Sheephead) Scientific Name: Semicossyphus pulcher Range: Sheephead range from the Gulf of California to Monterey Bay, California, although they are uncommon north of Point Conception. Habitat: Both adult and juvenile Sheephead primarily reside in kelp forest and rocky reef habitats. Size (length and weight): Male Sheephead can grow up to a length of three feet (91 centimeters) and weigh over 36 pounds (16 kilograms). Life span: The oldest Sheephead ever reported was a male at 53 years. Reproduction: As protogynous hermaphrodites, all Sheephead begin life as females, and older, larger females can develop into males. They are batch spawners, releasing eggs and sperm into the water column multiple times during their spawning season from July to September. Prey: Sheephead are generalist carnivores whose diet shifts throughout their growth. Juveniles primarily consume small invertebrates like tube-dwelling polychaetes, bryozoans and brittle stars, and adults shift to consuming larger mobile invertebrates like sea urchins. Predators: Predators of adult Sheephead include Giant Sea Bass, Soupfin Sharks and California Sea Lions. Fishery: Sheephead support both a popular recreational and a commercial fishery in southern California. Area fished: Sheephead are fished primarily south of Point Conception (Santa Barbara County) in nearshore waters around the offshore islands and along the mainland shore over rocky reefs and in kelp forests. Fishing season: Recreational anglers can fish for Sheephead from March 1through December 31 onboard boats south of Point Conception and May 1 through December 31 between Pigeon Point and Point Conception. Recreational divers and shore-based anglers can fish for Sheephead year round.