BGRS\SB-2018 MM&Am
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

RESULTS Long Jump Women - Heptathlon
Moscow (RUS) World Championships 10-18 August 2013 RESULTS Long Jump Women - Heptathlon RESULT NAME COUNTRY AGE DATE VENUE World Record 7291 Jackie JOYNER-KERSEE USA 26 24 Sep 1988 Seoul Championships Record 7128 Jackie JOYNER-KERSEE USA 25 1 Sep 1987 Roma World Leading 6623 Tatyana CHERNOVA RUS 25 8 Jul 2013 Kazan TEMPERATURE HUMIDITY START TIME 09:30 16° C 88 % Group A 13 August 2013 END TIME 10:24 19° C 77 % PLACE BIB NAME COUNTRY DATE of BIRTH ORDER RESULT POINTS 1 2 3 1 432 Claudia RATH GER 25 Apr 86 16 6.67 PB 6.67 6.44 X 1.0 m/s 1.0 m/s 0.4 m/s 1.0 m/s Клаудиа Рат 25 апр . 86 1062 2 391 Katarina JOHNSON-THOMPSON GBR 09 Jan 93 15 6.56 PB X 6.56 X 0.6 m/s 1.1 m/s 0.6 m/s 0.8 m/s Катарина Джонсон -Томпсон 09 янв . 93 1027 3 856 Ganna MELNICHENKO UKR 24 Apr 83 12 6.49 SB 6.23 6.49 X 0.2 m/s 0.3 m/s 0.2 m/s 0.3 m/s Анна Мельниченко 24 апр . 83 1004 4 203 Brianne THEISEN EATON CAN 18 Dec 88 8 6.37 PB X 6.37 X 0.5 m/s 1.0 m/s 0.5 m/s 1.3 m/s Брайанн Тайсен Итон 18 дек . 88 965 5 624 Dafne SCHIPPERS NED 15 Jun 92 14 6.35 6.35 6.14 6.14 1.3 m/s 1.3 m/s 0.6 m/s 0.7 m/s Дафна Шипперс 15 июня 92 959 6 672 Karolina TYMINSKA POL 04 Oct 84 13 6.32 6.32 X 6.16 0.9 m/s 0.9 m/s 0.7 m/s 0.7 m/s Каролина Тыминская 04 окт . -

Music Performance and Education
SCIENTIFIC JOURNAL OF MUSIC ART AND EDUCATION WORLD BULLETIN “MUSICAL ARTS AND EDUCATION OF THE UNESCO CHAIR IN LIFE-LONG LEARNING” The journal was founded in 2013 comes out 4 times a year Moscow State Pedagogical University was founded 140 years ago Founder: Federal State Budget Educational Institution of Higher Professional Education “Moscow State Pedagogical University” Publisher: “MSPU” Publishing House The co-publisher of the issue: FSBEIHPE “Mordovian State Pedagogical Institute named after M. E. Evsevyev” (branch of the UNESCO Chair “Musical Arts and Education in Life-long Learning” hosted by MordSPU) EDITORIAL BOARD A. L. Semenov (chairman of the board) Academician of Russian Academy of Sciences, Academician of Russian Academy of Education, Professor E. B. Abdullin (vice-chairman of the board) Professor, Doctor of Pedagogical Sciences, Member of the Union of Composers of Russia V. A. Gergiev People's Artist of Russia V. A. Gurevich Professor, Doctor of Arts, Secretary of the Union of Composers of Saint-Petersburg V. V. Kadakin Candidate of Pedagogical Sciences, Honored Worker of Education of the Republic of Mordovia B. M. Nemensky Professor, Academician of the Russian Academy of Education, People's Artist of Russia M. I. Roytershteyn Professor, Composer, Honoured Art Worker of Russia, Candidate of Arts A. S. Sokolov Professor, Doctor of Arts, Member of the Union of Composers of Russia G. M. Tsypin Professor, Doctor of Pedagogical Sciences, Candidate of Arts, Member of the Union of Composers of Russia R. K. Shedrin Composer, People's Artist of the USSR S. B. Yakovenko Professor, Doctor of Arts, People's Artist of Russia EDITORIAL BOARD Nikolaeva E. -

START LIST 100 Metres Hurdles Women - Heptathlon
REVISED Moscow (RUS) World Championships 10-18 August 2013 START LIST 100 Metres Hurdles Women - Heptathlon CONFIRMATION OF ATHLETES 377 MADE IN ERROR. REMOVE FROM START LIST. RESULT NAME COUNTRY AGE DATE VENUE World Record 7291 Jackie JOYNER-KERSEE USA 26 24 Sep 1988 Seoul Championships Record 7128 Jackie JOYNER-KERSEE USA 25 1 Sep 1987 Roma World Leading 6623 Tatyana CHERNOVA RUS 25 8 Jul 2013 Kazan i = Indoor performance Heat 1 12 August 2013 9:30 4 LANE BIB NAME COUNTRY DATE of BIRTH PERSONAL BEST SEASON BEST 2 321 Mari KLAUP EST 27 Feb 90 14.09 14.09 Мари Клауп 27 февр . 90 3 423 Julia MÄCHTIG GER 01 Jan 86 14.10 14.12 Юлия Мехтих 01 янв . 86 4 453 Györgyi FARKAS-ZSIVOCZKY HUN 13 Feb 85 13.97 13.97 Дьордьи Фаркаш -Живоцки 13 февр . 85 5 821 Wassana WINATHO THA 30 Jun 80 13.62 13.90 Вассана Уинато 30 июня 80 6 293 Eliška KLUCINOVÁ CZE 14 Apr 88 14.01 14.06 Элишка Клюцинова 14 апр . 88 7 640 Ida MARCUSSEN NOR 01 Nov 87 13.96 14.11 Ида Маркуссен 01 нояб . 87 8 713 Aleksandra BUTVINA RUS 14 Feb 86 14.04 14.04 Александра Бутвина 14 февр . 86 9 545 Irina KARPOVA KAZ 13 Feb 80 13.96 14.52 Ирина Карпова 13 февр . 80 Heat 2 12 August 2013 9:38 4 LANE BIB NAME COUNTRY DATE of BIRTH PERSONAL BEST SEASON BEST 2 163 Katsiaryna NETSVIATAYEVA BLR 28 Jun 89 13.74 13.84 Катерина Нецвятаева 28 июня 89 3 804 Linda ZÜBLIN SUI 21 Mar 86 13.55 13.76 Линда Зюблин 21 марта 86 4 148 Nafissatou THIAM BEL 19 Aug 94 13.87 13.87 Нафиссату Тиам 19 авг . -

Peoples, Identities and Regions. Spain, Russia and the Challenges of the Multi-Ethnic State
Institute Ethnology and Anthropology Russian Academy of Sciences Peoples, Identities and Regions. Spain, Russia and the Challenges of the Multi-Ethnic State Moscow 2015 ББК 63.5 УДК 394+312+316 P41 Peoples, Identities and Regions. Spain, Russia and the Challenges of the Multi-Ethnic State / P41 edited by Marina Martynova, David Peterson, Ro- man Ignatiev & Nerea Madariaga. Moscow: IEA RAS, 2015. – 377 p. ISBN 978-5-4211-0136-9 This book marks the beginning of a new phase in what we hope will be a fruitful collaboration between the Institute Ethnology and Anthropology Russian Academy of Science and the University of the Basque Country. Researchers from both Spain and Russia, representing a series of scientific schools each with its own methods and concepts – among them anthropologists, political scien- tists, historians and literary critics-, came to the decision to prepare a collective volume exploring a series of vital issues concerning state policy in complex societies, examining different identitarian characteristics, and reflecting on the difficulty of preserving regional cultures. Though the two countries clearly have their differences – political, economic and social –, we believe that the compar- ative methodology and the debates it leads to are valid and indeed important not just at a theoretical level, but also in practical terms. The decision to publish the volume in English is precisely to enable us to overcome any linguistic barriers there might be between Russian and Spanish academics, whilst simultaneously making these studies accessible to a much wider audience, since the realities be- hind many of the themes touched upon in this volume are relevant in many other parts of the globe beyond our two countries. -

63Rd Annual Meeting Program.Pdf
Program of the 63rd Annual Meeting of the International Society of Electrochemistry iii The 63rd Annual Meeting of the International Society of Electrochemistry Electrochemistry for Advanced Materials, Technologies and Instrumentation 19-24 August, 2012, Prague, Czech Republic Contents list Organizing Committee ...................................................................................................................v Symposium Organizers ...........................................................................................................vi-vii Tutorial Lectures ........................................................................................................................ viii Plenary Lectures ............................................................................................................................ix Prize Winners ............................................................................................................................ x-xi Special Meetings ......................................................................................................................... xii Overall Schedule by Day ........................................................................................................... xiii Poster Sessions ........................................................................................................................... xiii General Information ....................................................................................................................xiv -

London 2012 Athletics Day 1 Results
http://www.iaaf.org/results/eventCode=4871/raceDate=08-03-2012/txt... ------------------------------------------------------------------------------------- The XXX Olympic Games London (OS) (GBR) - Friday, Aug 03, 2012 Shot Put - M QUALIFICATION Qual. rule: qualification standard 20.65m or at least best 12 qualified. ------------------------------------------------------------------------------------- Group A 03 August 2012 - 10:00 Position Bib Athlete Country Mark . 1 1924 David Storl GER 21.15 Q . 2 2659 Tomasz Majewski POL 21.03 Q . 3 1023 Germán Lauro ARG 20.75 Q (NR) 4 1163 Pavel Lyzhyn BLR 20.57 q (SB) 5 3209 Christian Cantwell USA 20.41 q . 6 2187 Dorian Scott JAM 20.31 q . 7 2836 Soslan Tsirikhov RUS 20.17 . 8 2550 Rutger Smith NED 20.08 . 9 2711 Marco Fortes POR 20.06 . 10 2046 Lajos Kürthy HUN 19.65 . 11 1264 Georgi Ivanov BUL 19.63 . 12 1153 Kemal Mesic BIH 19.60 . 13 1986 Mihaíl Stamatóyiannis GRE 19.24 . 14 1063 Dale Stevenson AUS 19.17 . 15 2395 Maris Urtans LAT 19.13 . 16 1639 Borja Vivas ESP 18.88 . 17 2505 Stephen Saenz MEX 18.65 . 18 2118 Ódinn Björn Thorsteinsson ISL 17.62 . 19 1003 Adriatik Hoxha ALB 17.58 . 1297 Justin Rodhe CAN NM . Athlete 1st 2nd 3rd David Storl 21.15 Tomasz Majewski 21.03 Germán Lauro 20.10 X 20.75 Pavel Lyzhyn 19.85 20.10 20.57 Christian Cantwell 20.41 20.30 19.88 Dorian Scott 20.18 20.30 20.31 Soslan Tsirikhov 19.25 19.78 20.17 Rutger Smith 20.08 X 19.55 Marco Fortes 19.59 20.06 19.50 Lajos Kürthy 19.65 X X Georgi Ivanov 19.42 19.40 19.63 Kemal Mesic 19.49 X 19.60 Mihaíl Stamatóyiannis 18.67 X 19.24 Dale Stevenson 18.38 19.17 19.01 Maris Urtans 18.92 19.13 X Borja Vivas 18.46 X 18.88 Stephen Saenz X 18.65 X Ódinn Björn Thorsteinsson X 17.04 17.62 Adriatik Hoxha 17.58 X 17.13 Justin Rodhe X X X Group B 03 August 2012 - 10:00 Position Bib Athlete Country Mark . -
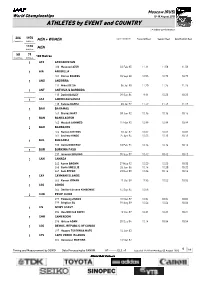
0 E Event Country
Moscow (RUS) World Championships 10-18 August 2013 ATHLETES by EVENT and COUNTRY i = Indoor performance 206 1970 MEN + WOMEN DATE of BIRTH Personal Best Season Best Qualification Best Countries Athletes 1105 MEN Athletes 58 79 100 Metres Countries Athletes 1 AFG AFGHANISTAN 100 Massoud AZIZI 02 Feb 85 11.11 11.58 11.58 1 AIA ANGUILLA 101 Kieron ROGERS 08 Sep 88 10.55 10.75 10.75 1 AND ANDORRA 111 Mikel DE SA 06 Jul 90 11.20 11.26 11.26 1 ANT ANTIGUA & BARBUDA 113 Daniel BAILEY 09 Sep 86 9.91 10.23 10.23 1 ASA AMERICAN SAMOA 118 Faresa KAPISI 23 Jul 97 11.47 11.47 11.47 1 BAH BAHAMAS 161 Shavez HART 09 Jun 92 10.16 10.16 10.16 1 BAN BANGLADESH 165 Masbah AHMMED 11 Mar 95 10.99 10.99 10.99 2 BAR BARBADOS 166 Ramon GITTENS 20 Jul 87 10.02 10.02 10.02 167 Andrew HINDS 25 Apr 84 10.03 10.13 10.13 1 BUL BULGARIA 233 Denis DIMITROV 10 Feb 94 10.16 10.16 10.16 1 BUR BURKINA FASO 237 Innocent BOLOGO 05 Sep 89 10.42 10.42 10.42 3 CAN CANADA 243 Aaron BROWN 27 May 92 10.05 10.05 10.05 245 Gavin SMELLIE 26 Jun 86 10.14 10.20 10.20 247 Sam EFFAH 29 Dec 88 10.06 10.14 10.14 1 CAY CAYMAN ISLANDS 263 Kemar HYMAN 11 Oct 89 9.95 10.02 10.02 1 CGO CONGO 264 Devilert Arsene KIMBEMBE 14 Sep 84 10.58 2 CHN PR OF CHINA 277 Peimeng ZHANG 13 Mar 87 10.04 10.04 10.04 279 Bingtian SU 29 Aug 89 10.06 10.06 10.06 1 CIV IVORY COAST 294 Hua Wilfried KOFFI 12 Oct 87 10.21 10.21 10.21 1 CMR CAMEROON 296 Idrissa ADAM 28 Dec 84 10.14 10.54 10.54 1 COD DEMOC. -

20Th Asian Athletics Championships
20th Asian Athletics Championships-2013 * 03rd to 07th July 2013 * Pune, Maharastra, INDIA * *~*~* Final Results *~*~* Event # 1 MENS 100m Time:-19:15 Hrs 04/07/2013 # W.R.:-:USAIN BOLT JAM,BERLIN,GER 9.58 16-8-2009 % A.R.:-:Samuel Francis QAT;Amman 9.99 26.07.2007 &M.R.:-:S.Francis QAT; Amman 9.99 26.07.2007 Final 1 SU BINGTIAN 29/08/89 CHN -0.3m/s 10.17 2 SAMUEL ADELEBARI FRANCIS 27/03/87 QAT -0.3m/s 10.27 3 BARAKAT AL HARTHI 15/06/88 OMA -0.3m/s 10.30 4 REZA GHASEMI 24/07/87 IRI -0.3m/s 10.37 5 FAHAD MOHAMMED ALSUBAIE 04/02/94 KSA -0.3m/s 10.48 6 NG KA FUNG 27/10/92 HKG -0.3m/s 10.52 7 NAOKI TSUKAHARA 10/05/85 JPN -0.3m/s 10.54 8 HASSAN TAFTIAN 04/05/93 IRI -0.3m/s 10.54 Semi Final-1 1 SU BINGTIAN 29/08/89 CHN -1.1M/S 10.37 2 REZA GHASEMI 24/07/87 IRI -1.1M/S 10.38 3 FAHAD MOHAMMED ALSUBAIE 04/02/94 KSA -1.1M/S 10.48 4 TSUI CHI HO 17/02/90 HKG -1.1M/S 10.52 5 ANIRUDDH KALIDAS GUJJAR 14/11/90 IND -1.1M/S 10.59 6 MOHAMED SALMAN 04/04/92 BRN -1.1M/S 10.86 EID ABDULLA E A AL KUWARI 04/02/93 QAT DQ SOTA KAWATSURA 19/06/89 JPN DQ Semi Final-2 1 BARAKAT AL HARTHI 15/06/88 OMA -0.5M/S 10.27 2 SAMUEL ADELEBARI FRANCIS 27/03/87 QAT -0.5M/S 10.30 3 HASSAN TAFTIAN 04/05/93 IRI -0.5M/S 10.32 4 NAOKI TSUKAHARA 10/05/85 JPN -0.5M/S 10.44 5 NG KA FUNG 27/10/92 HKG -0.5M/S 10.47 6 ALI AL DOSERI 02/03/83 BRN -0.5M/S 10.59 7 LIAQAT ALI 06/08/83 PAK -0.5M/S 10.64 8 SAPWATUR RAHMAN 13/05/94 INA -0.5M/S 10.78 Heat-1 1 REZA GHASEMI 24/07/87 IRI -0.2m/s 10.41 2 NG KA FUNG 27/10/92 HKG -0.2m/s 10.51 3 ALI AL DOSERI 02/03/83 BRN -0.2m/s 10.72 4 KRITSADA NAMSUWAN -

ETN2012 53(Wogii)
Volume 11, No. 53 August 17, 2012 version ii — Olympic Games Women — — By–Nation Medal Chart — LONDON, ENGLAND AUGUST 3–12 Nation ................Men Women Overall G S B Total G S B Total ATTENDANCE United States ......3 9 3 15 6 4 4 14 ........29 No official figures released, but all sessions should be considered Russia .................2 — 2 6 5 5 16 ........18 “80,000-seat sellouts.” Jamaica ..............3 2 2 7 1 2 2 5 ..........12 WEATHER Kenya ..................2 1 4 7 — 3 1 4 ..........11 Official temperature (both F and C) and humidity readings are given Germany .............1 2 1 4 — 2 2 4 ............8 with each event. Spotty rain on several days; generally pleasant Ethiopia ...............— 1 1 2 3 — 2 5 ............7 (c70F/20C) and calm. In almost all cases the readings are for the China ..................1 — 2 3 — — 3 3 ............6 beginning of the event. Great Britain ........3 — 1 4 1 1 — 2 ............6 Trinidad ...............1 — 3 4 — — — — ...........4 EXPLAINING THE TYPOGRAPHY Paragraph breaks in the preliminary rounds of running events indicate Australia ..............— 2 — 2 1 — — 1 ............3 the separation between qualifiers and non-qualifiers. Ukraine ...............— 1 — 1 — — 2 2 ............3 Cuba ...................— 1 1 — 1 — 1 ............2 COLOR CODING Czech Republic...— — — 1 — 1 2 ............2 All medalists appear in purple ink; all Americans are underlined (if Dominican Rep ...1 1 — 2 — — — — ...........2 in multiple rounds, only in the final round in which they competed); France .................1 1 — 2 — — — — ...........2 field-event/multi medalists appear in either blue (gold medal), red Poland ................1 — 1 — 1 — 1 ............2 (silver) or green (bronze) in the progression charts. -
Asian Athletics Digest February 15, 2016 4 / 2016
Asian Athletics Digest February 15, 2016 4 / 2016 Asian Indoor Championships Special Preview ~ Ranklists ~ Records Asian Athletics Digest February 15, 2016 Contents From the Editor’s desk .……………… 66 Preview on Asian Indoor Champs …. 67 From the Editor’s desk Asian Indoor All-Time Rankings ……. 68 Asian & National Indoor Records ….. 75 Hello All, Welcome back to Asian Athletics Digest! First of all I would like to thank you all for your No. 4/2016 comments, continuous support and encouragement to our magazine from time to time. Editor & Publisher Ram. Murali Krishnan This edition of our magazine is dedicated to the 7th Asian Indoor Athletics Championships to be held Doha, Qatar, from 19 Address for Communication to 20 February 2016. “ Kausalyam ” 72/4, Sri Padmavathi Nagar NSN Palayam P.O. As in the previous edition, that was produced as a Coimbatore – 641031, INDIA special preview for the South Asian Games, this number of our magazine will also have tons of stats on Asian Indoor Athletics Contact E-Mail [email protected] As much as 432 athletes from 37 member national [email protected] federations of Asian Athletics Association are expected to take part in the three day meet at Doha. PHOTOS Doha 2016 Organising Committee In this edition of Asian Athletics Digest contains a brief history of Asian Indoor championships, Asian All-Time Top-10 Designer ranking lists as well as Asian and National Indoor Records. N. Sasivarnan To accommodate the large number of info in this issue we have excluded some of our regular features, which will be resumed in our next number. -

Alexander Bolonkin
1 Alexander Bolonkin USSR NASA, USA LIFE. SCIENCE. FUTURE Concentration camp, USSR. New York 2 Alexander Bolonkin LIFE. SCIENCE. FUTURE (Biography notes, researches and innovations. Translation from Russian by Yulia Plotnikov) New York 3 Acknowledgement The author thanks Professor at the University of Perm Oleg Pensky for active endpoint support the publication of this book, Julia Plotnikov for translation and explanation of abbreviations. 4 Contents Preface 6 Part 1. Life in the USSR 7 1. Ancestors.(from the memoirs of sister Anne Bolonkin). 7 2. Childhood 10 3. Perm Aviation College and the Regional Laboratory of model aircraft (1948-52) 15 4. Kazan Aviation Institute (KAI, 1952-1958) 19 5. Experimental Design Bureau of O.K. Antonov 27 6. Doctoral Dissertation. Moscow Aviation Institute. 37 7. Experimental Design Bureau of V.P. Glushko 40 8. Moon Race 48 9. Moscow Aviation Technology Institute (MATI). Moscow Higher Technical University named Bauman (MHTU) 61 10. Post-Doctoral dissertation 62 Part2. In Soviet Concentration Camps 67 1. Preface 68 2. Acceding to remedial activity 68 3. Propaganda leaflets 73 4. Arrest, investigation 73 5. Trial 76 6. Concentration camp JH-389/17а 77 7. Concentration camp hospital 78 8. Concentration camp JH-389/19 82 9. Penalty isolation ward and penalty cell (special prison) 84 10. Concentration camp in Barashevo 85 11. Road to exile 89 12. First exile 90 13. Second arrest 98 14. Ulan-Ude prison 99 15. Second trial. Concentration camp ОV-94/2 102 16. Third arrest and fabrication of a damning ―new case‖ 105 17. Second exile. 107 APPENDIX #1 according to the materials of radio station ―Svoboda‖/(―Freedom‖) 111. -

Deep Impact: Robert Goddard and the Soviet 'Space Fad' of the 1920S
History and Technology Vol. 20, No. 2, June 2004, pp. 97–113 Deep Impact: Robert Goddard and the Soviet ‘Space Fad’ of the 1920s Asif Siddiqi UsingTaylorghat041008.sgm10.1080/0734151042000240782History0734-1512Original2004202000000JuneAsifSiddiqiasiddiqi@andrew.cmu.edu and& and Article Francis (print)Francis Technology2004 Ltd Ltd newly available information from Russian archives, this paper explores American rocketry pioneer Robert Goddard’s relationship to the Soviet space-flight advocacy commu- nity in the 1920s. In post-Revolutionary Russia, Goddard enjoyed a curious kind of fame. News of his alleged plan to launch a rocket to the Moon permeated widely through a Soviet audience interested in the possibility of space exploration. Goddard’s practical work in developing rockets became a metaphor for the aspirations of the many in Soviet Russia who were unwilling to limit their horizons to theory and prognostication. The new research into Goddard’s relationship to the Soviet space-flight enthusiast community underscores how international contacts shaped the space advocacy movements of the early twentieth century. The new evidence prompts us to consider an alternative approach to the ‘founda- tion myth’ of space history involving Tsiolkovskii, Goddard and Oberth, one that privileges an international context instead of the usual multiple national contexts. Keywords: Robert Goddard; Soviet Union; America; Spaceflight; Popularization; 1920s Introduction In the history of astronautics, it has become a cliché to begin all narratives by invoking the Russian Konstantin Tsiolkovskii, the Romanian (and later German) Hermann Oberth and the American Robert Goddard. In the early decades of the twentieth century, the three men independently produced the first serious theoretical works on the possibility of rocketry and space travel.