MBN Explorer Users' Guide
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Modeling the Effect of Ion-Induced Shock Waves and DNA Breakage
Modeling the effect of ion-induced shock waves and DNA breakage with the reactive CHARMM force field Ida Friis,1 Alexey V. Verkhovtsev,2, ∗ Ilia A. Solov’yov,3, ∗ and Andrey V. Solov’yov2, ∗ 1Department of Physics, Chemistry and Pharmacy, University of Southern Denmark, Campusvej 55, 5230 Odense M, Denmark 2MBN Research Center, Altenh¨oferallee 3, 60438 Frankfurt am Main, Germany 3Department of Physics, Carl von Ossietzky Universit¨at Oldenburg, Carl-von-Ossietzky-Str. 9-11, 26129 Oldenburg, Germany Ion-induced DNA damage is an important effect underlying ion beam cancer therapy. This paper introduces the methodology of modeling DNA damage induced by a shock wave caused by a projectile ion. Specifically it is demonstrated how single- and double strand breaks in a DNA molecule could be described by the reactive CHARMM (rCHARMM) force field implemented in the program MBN Explorer. The entire work flow of performing the shock wave simulations, including obtaining the crucial simulation parameters, is described in seven steps. Two exemplary analyses are provided for a case study simulation serving to: (i) quantify the shock wave propagation, and (ii) describe the dynamics of formation of DNA breaks. The paper concludes by discussing the computational cost of the simulations and revealing the possible maximal computational time for different simulation setups. I. INTRODUCTION changes of their topology and molecular composition [9, 11, 12]. It takes into account additional parameters Classical molecular dynamics (MD) enables studying of the system, such as bond dissociation energy, bond of structure and dynamics of biological systems and inor- order and the valence of atoms. -
![Arxiv:1701.02495V2 [Cond-Mat.Mtrl-Sci] 6 Jul 2017](https://docslib.b-cdn.net/cover/0023/arxiv-1701-02495v2-cond-mat-mtrl-sci-6-jul-2017-8010023.webp)
Arxiv:1701.02495V2 [Cond-Mat.Mtrl-Sci] 6 Jul 2017
ATK-ForceField: A New Generation Molecular Dynamics Software Package Julian Schneider1, Jan Hamaekers2, Samuel T. Chill1, Søren Smidstrup1, Johannes Bulin2, Ralph Thesen2, Anders Blom1, Kurt Stokbro1 1 QuantumWise A/S, Fruebjergvej 3, DK-2100 Copenhagen, Denmark 2 Fraunhofer Institute for Algorithms and Scientific Computing SCAI, Schloss Birlinghoven, 53754 Sankt Augustin, Germany E-mail: [email protected] Abstract. ATK-ForceField is a software package for atomistic simulations using classical interatomic potentials. It is implemented as a part of the Atomistix ToolKit (ATK), which is a Python programming environment that makes it easy to create and analyze both standard and highly customized simulations. This paper will focus on the atomic interaction potentials, molecular dynamics, and geometry optimization features of the software, however, many more advanced modeling features are available. The implementation details of these algorithms and their computational performance will be shown. We present three illustrative examples of the types of calculations that are possible with ATK-ForceField: modeling thermal transport properties in a silicon germanium crystal, vapor deposition of selenium molecules on a selenium surface, and a simulation of creep in a copper polycrystal. 1. Introduction Molecular dynamics (MD) simulations have become a versatile and widely used tool in scientific computer simulations [1, 2]. MD can be used to gain insight in atomic-scale processes when experimental techniques are unable to provide sufficient resolution. MD is used to sample microscopic ensembles, to study microscopic dynamical and transport properties, such as diffusion or thermal transport, or to simulate physical processes in order to access atomic structures for which little experimental information is available, e.g. -

Research Report Hochleistungsrechner in Hessen 2010/2011 Inhalt
Research Report Hochleistungsrechner in Hessen 2010/2011 Inhalt Vorwort zum Jahresbericht 2010-2011 ......................................................................................................................................................................................... 7 Die Hochleistungsrechner Der Hessische Hochleistungsrechner an der Technischen Universität Darmstadt ...................................................................................................8 Die Hochleistungsrechner (HLR) am Center for Scientific Computing (CSC) der Goethe-Universität ..................................................10 Anpassung des LINPACK-Benchmarks für den LOEWE-CSC ............................................................................................................................................12 Das Linux-Cluster im IT-Servicezentrum der Universität Kassel ........................................................................................................................................13 Die Hochleistungsrechner der GSI ........................................................................................................................................................................................................14 Die Rechencluster der Philipps-Universität Marburg.................................................................................................................................................................16 Das zentrale Hochleistungsrechen-Cluster der Justus-Liebig-Universität Gießen -
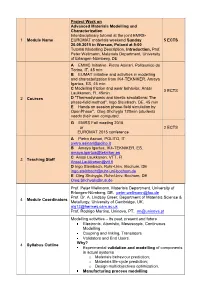
Advanced Materials Modelling and Characterization.Pdf
Project Work on Advanced Materials Modelling and Characterization Interdisciplinary tutorial at the joint EMRS- 1 Module Name EUROMAT materials weekend Sunday 5 ECTS 20.09.2015 in Warsaw, Poland at 9:00 Tutorial Modelling Description, Introduction, Prof. Peter Wellmann, Materials Department, University of Erlangen-Nürnberg, DE A EMMC Initiative, Pietro Asinari, Politecnico do Torino, IT, 45 min B EUMAT Initiative and activities in modelling and characterization from IK4-TEKNIKER, Amaya Igartua, ES, 45 min C Modelling friction and wear behavior, Anssi 3 ECTS Laukkanen, FI, 45min 2 Courses D "Thermodynamic and kinetic simulations: The phase-field method", Ingo Steinbach, DE, 45 min E Hands on session phase-field simulation by OpenPhase", Oleg Shchyglo 120min (students needs their own computer). G EMRS Fall meeting 2015 or 2 ECTS EUROMAT 2015 conference A Pietro Asinari, POLITO, IT [email protected] B Amaya Igartua, IK4-TEKNIKER, ES, [email protected] C Anssi Laukkanen, VTT, FI 3 Teaching Staff [email protected] D Ingo Steinbach, Ruhr-Univ. Bochum, DE [email protected] E Oleg Shchyglo, Ruhr-Univ. Bochum, DE [email protected] Prof. Peter Wellmann, Materials Department, University of Erlangen-Nürnberg, DE, [email protected] Prof. Dr. A. Lindsay Greer, Department of Materials Science & 4 Module Coordinators Metallurgy, University of Cambridge, UK, [email protected] Prof. Rodrigo Martins, Uninova, PT, [email protected] Modelling activities – its past, present and future Electronic, Atomistic, Mesoscopic, Continuous Modelling Coupling and linking, Translators Validators and End Users. Why? 4 Syllabus Outline Experimental validation and modelling of components in actual systems o Materials behaviour prediction; o Materials life-cycle prediction; o Design multiobjectives optimization. -

MBN Explorer Users' Guide Describes How to Run and to Use the Various Features of the Multiscale Program MBN Explorer
A universal program for multiscale simulation of complex molecular structure and dynamics MBN Explorer Users’ Guide Version 3.0 (March 31, 2017) Current editors: Ilia A. Solov’yov, Gennady Sushko and Andrey V. Solov’yov URL: www.mbnexplorer.com E-mail: [email protected] Description The MBN Explorer Users' Guide describes how to run and to use the various features of the multiscale program MBN Explorer. This guide includes the descrip- tion of the main features of the program, the manual how to use the program, the specication of input les and formats, and instructions on how to run the program on Windows, Linux/Unix and Macintosh platforms. Contents 1 Introduction 1 1.1 Features of MBN Explorer ..................... 1 1.2 MBN Explorer functionality .................... 3 1.3 Historical remarks ............................ 5 1.4 Terms and conditions .......................... 9 1.5 Third party terms and conditions ................... 18 1.5.1 VMD molle plugins ...................... 18 1.5.2 Mersenne twister ........................ 19 1.5.3 Kiss FFT ............................ 19 2 Getting Started 21 2.1 What is needed ............................. 21 2.2 MBN Explorer task le ....................... 21 2.2.1 Task le syntax ......................... 22 2.2.2 Required parameters ...................... 22 2.3 MBN Explorer task parameters . 22 2.3.1 General parameters ....................... 22 2.3.2 Input les ............................ 24 2.3.3 Output les and related parameters . 25 2.3.4 General numerical methods for interatomic interactions . 30 2.3.5 Ewald summation method for Coulomb interactions . 33 2.3.6 Fast particle mesh Ewald algorithm . 34 2.3.7 General molecular dynamics simulation . 36 2.3.8 Temperature control .....................