Oct 2020 Blood Group Typing from Whole-Genome Sequencing Data J Paganini, PL Nagy, N Rouse
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene List HTG Edgeseq Immuno-Oncology Assay
Gene List HTG EdgeSeq Immuno-Oncology Assay Adhesion ADGRE5 CLEC4A CLEC7A IBSP ICAM4 ITGA5 ITGB1 L1CAM MBL2 SELE ALCAM CLEC4C DST ICAM1 ITGA1 ITGA6 ITGB2 LGALS1 MUC1 SVIL CDH1 CLEC5A EPCAM ICAM2 ITGA2 ITGAL ITGB3 LGALS3 NCAM1 THBS1 CDH5 CLEC6A FN1 ICAM3 ITGA4 ITGAM ITGB4 LGALS9 PVR THY1 Apoptosis APAF1 BCL2 BID CARD11 CASP10 CASP8 FADD NOD1 SSX1 TP53 TRAF3 BCL10 BCL2L1 BIRC5 CASP1 CASP3 DDX58 NLRP3 NOD2 TIMP1 TRAF2 TRAF6 B-Cell Function BLNK BTLA CD22 CD79A FAS FCER2 IKBKG PAX5 SLAMF1 SLAMF7 SPN BTK CD19 CD24 EBF4 FASLG IKBKB MS4A1 RAG1 SLAMF6 SPI1 Cell Cycle ABL1 ATF1 ATM BATF CCND1 CDK1 CDKN1B NCL RELA SSX1 TBX21 TP53 ABL2 ATF2 AXL BAX CCND3 CDKN1A EGR1 REL RELB TBK1 TIMP1 TTK Cell Signaling ADORA2A DUSP4 HES1 IGF2R LYN MAPK1 MUC1 NOTCH1 RIPK2 SMAD3 STAT5B AKT3 DUSP6 HES5 IKZF1 MAF MAPK11 MYC PIK3CD RNF4 SOCS1 STAT6 BCL6 ELK1 HEY1 IKZF2 MAP2K1 MAPK14 NFATC1 PIK3CG RORC SOCS3 SYK CEBPB EP300 HEY2 IKZF3 MAP2K2 MAPK3 NFATC3 POU2F2 RUNX1 SPINK5 TAL1 CIITA ETS1 HEYL JAK1 MAP2K4 MAPK8 NFATC4 PRKCD RUNX3 STAT1 TCF7 CREB1 FLT3 HMGB1 JAK2 MAP2K7 MAPKAPK2 NFKB1 PRKCE S100B STAT2 TYK2 CREB5 FOS HRAS JAK3 MAP3K1 MEF2C NFKB2 PTEN SEMA4D STAT3 CREBBP GATA3 IGF1R KIT MAP3K5 MTDH NFKBIA PYCARD SMAD2 STAT4 Chemokine CCL1 CCL16 CCL20 CCL25 CCL4 CCR2 CCR7 CX3CL1 CXCL12 CXCL3 CXCR1 CXCR6 CCL11 CCL17 CCL21 CCL26 CCL5 CCR3 CCR9 CX3CR1 CXCL13 CXCL5 CXCR2 MST1R CCL13 CCL18 CCL22 CCL27 CCL7 CCR4 CCRL2 CXCL1 CXCL14 CXCL6 CXCR3 PPBP CCL14 CCL19 CCL23 CCL28 CCL8 CCR5 CKLF CXCL10 CXCL16 CXCL8 CXCR4 XCL2 CCL15 CCL2 CCL24 CCL3 CCR1 CCR6 CMKLR1 CXCL11 CXCL2 CXCL9 CXCR5 -

Rabbit Anti-Phospho-CD18-SL10462R-FITC
SunLong Biotech Co.,LTD Tel: 0086-571- 56623320 Fax:0086-571- 56623318 E-mail:[email protected] www.sunlongbiotech.com Rabbit Anti-Phospho-CD18 SL10462R-FITC Product Name: Anti-Phospho-CD18 (Thr758)/FITC Chinese Name: FITC标记的磷酸化整合素β2/Integrin β2抗体 CD18 (Phospho Thr758); CD18 (Phospho-Thr758); CD18 (Phospho T758); p-CD18 (T758); p-CD18 (Thr758); Integrin beta 2; 95 subunit beta; CD 18; CD18; Cell surface adhesion glycoprotein LFA 1/CR3/P150,959 beta subunit precursor); Cell surface adhesion glycoproteins LFA 1/CR3/p150,95 subunit beta; Cell surface adhesion glycoproteins LFA-1/CR3/p150; Complement receptor C3 beta subunit; Complement Alias: receptor C3 subunit beta; Integrin beta chain beta 2; Integrin beta-2; Integrin, beta 2 (complement component 3 receptor 3 and 4 subunit); ITB2_HUMAN; ITGB2; LAD; LCAMB; Leukocyte associated antigens CD18/11A, CD18/11B, CD18/11C; Leukocyte cell adhesion molecule CD18; LFA 1; LFA1; Lymphocyte function associated antigen 1; MAC 1; MAC1; MF17; MFI7; OTTHUMP00000115278; OTTHUMP00000115279; OTTHUMP00000115280; OTTHUMP00000115281; OTTHUMP00000115282. Organism Species: Rabbit Clonality: Polyclonal React Species: Human,Mouse,Rat,Chicken,Dog,Pig,Cow,Horse,Rabbit,Sheep,Guinea Pig, Flow-Cyt=1:50-200ICC=1:50-200IF=1:50-200www.sunlongbiotech.com Applications: not yet tested in other applications. optimal dilutions/concentrations should be determined by the end user. Molecular weight: 82kDa Form: Lyophilized or Liquid Concentration: 1mg/ml KLH conjugated synthesised phosphopeptide derived from human CD18 around the immunogen: phosphorylation site of Thr758 Lsotype: IgG Purification: affinity purified by Protein A Storage Buffer: 0.01M TBS(pH7.4) with 1% BSA, 0.03% Proclin300 and 50% Glycerol. Store at -20 °C for one year. -

Anti-ICAM4 Monoclonal Antibody (DCABH-11966) This Product Is for Research Use Only and Is Not Intended for Diagnostic Use
Anti-ICAM4 monoclonal antibody (DCABH-11966) This product is for research use only and is not intended for diagnostic use. PRODUCT INFORMATION Antigen Description This gene encodes the Landsteiner-Wiener (LW) blood group antigen(s) that belongs to the immunoglobulin (Ig) superfamily, and that shares similarity with the intercellular adhesion molecule (ICAM) protein family. This ICAM protein contains 2 Ig-like C2-type domains and binds to the leukocyte adhesion LFA-1 protein. The molecular basis of the LW(A)/LW(B) blood group antigens is a single aa variation at position 100; Gln-100=LW(A) and Arg-100=LW(B). Alternative splicing results in multiple transcript variants encoding distinct isoforms. Immunogen A synthetic peptide of human ICAM4 is used for rabbit immunization. Isotype IgG Source/Host Rabbit Species Reactivity Human Purification Protein A Conjugate Unconjugated Applications Western Blot (Transfected lysate); ELISA Size 1 ea Buffer In 1x PBS, pH 7.4 Preservative None Storage Store at -20°C or lower. Aliquot to avoid repeated freezing and thawing. GENE INFORMATION Gene Name ICAM4 intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) [ Homo sapiens ] Official Symbol ICAM4 Synonyms ICAM4; intercellular adhesion molecule 4 (Landsteiner-Wiener blood group); intercellular adhesion molecule 4 (LW blood group) , intercellular adhesion molecule 4, Landsteiner Wiener blood group , Landsteiner Wiener blood group , LW; intercellular adhesion molecule 4; CD242; CD242 antigen; LW blood group protein; Landsteiner-Wiener blood group -

Genetic Testing Policy Number: PG0041 ADVANTAGE | ELITE | HMO Last Review: 04/11/2021
Genetic Testing Policy Number: PG0041 ADVANTAGE | ELITE | HMO Last Review: 04/11/2021 INDIVIDUAL MARKETPLACE | PROMEDICA MEDICARE PLAN | PPO GUIDELINES This policy does not certify benefits or authorization of benefits, which is designated by each individual policyholder terms, conditions, exclusions and limitations contract. It does not constitute a contract or guarantee regarding coverage or reimbursement/payment. Paramount applies coding edits to all medical claims through coding logic software to evaluate the accuracy and adherence to accepted national standards. This medical policy is solely for guiding medical necessity and explaining correct procedure reporting used to assist in making coverage decisions and administering benefits. SCOPE X Professional X Facility DESCRIPTION A genetic test is the analysis of human DNA, RNA, chromosomes, proteins, or certain metabolites in order to detect alterations related to a heritable or acquired disorder. This can be accomplished by directly examining the DNA or RNA that makes up a gene (direct testing), looking at markers co-inherited with a disease-causing gene (linkage testing), assaying certain metabolites (biochemical testing), or examining the chromosomes (cytogenetic testing). Clinical genetic tests are those in which specimens are examined and results reported to the provider or patient for the purpose of diagnosis, prevention or treatment in the care of individual patients. Genetic testing is performed for a variety of intended uses: Diagnostic testing (to diagnose disease) Predictive -

An Overview About Erythrocyte Membrane
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Universidade de Lisboa: Repositório.UL Clinical Hemorheology and Microcirculation 44 (2010) 63–74 63 DOI 10.3233/CH-2010-1253 IOS Press An overview about erythrocyte membrane Sofia de Oliveira ∗ and Carlota Saldanha 1 Unidade de Biologia Microvascular e Inflamação, Instituto de Medicina Molecular, Instituto de Bioquímica, Faculdade de Medicina, Universidade de Lisboa, Lisboa, Portugal Received 29 May 2009 Accepted 7 September 2009 Abstract. In the sixties and seventies, erythrocytes or red blood cells (RBCs) were extensively studied. Much has been learnt particularly concerning their metabolism and gas transporter function. In the past decade, the use of new approaches and methodologies, such as proteomic analysis, has contributed for a renewed interest on the erythrocyte. Recent studies have provided us with a more detailed and comprehensive picture on the composition and organization of its cellular membrane that will be the main subject of this minireview. Unexpectedly, it has been recognized that this cell expresses several adhesion molecules on its surface, like other cellular types such blood circulating cells or endothelial cells. Taking into consideration the cellular functions of the erythrocyte, the clarification of the role of those adhesion molecules may in the future open new horizons for the biological significance of this cellular player. Keywords: Erythrocyte, erythrocyte membrane, membrane structure and adhesion molecules 1. Introduction Erythrocytes are the major cell component of blood. These red cells are a product of a differentiation process that starts in the bone marrow where hematopoietic stem cells differentiate to nucleate RBCs. -

Red Blood Cell Antigen Genotyping
Red Blood Cell Antigen Genotyping Testing is useful in determining allelic variants predicting red blood cell (RBC) antigen phenotypes for patients with recent history of transfusion or with conflicting serological antibody results due to partial, variant, or weak expression antigens. Also Tests to Consider useful as an aid in management of hemolytic disease of the fetus and newborn (HDFN). Typical Testing Strategy Disease Overview Phenotype Testing Evaluates specific RBC antigen presence by serology Prevalence and/or Incidence Results can aid in selecting antigen negative RBC units Erythrocyte alloimmunization occurs in up to 58% of sickle cell patients, up to 35% in other transfusion-dependent patients, and in approximately 0.8% of all pregnant Antigen Testing, RBC Phenotype women. Extended 0013020 Method: Hemagglutination Serological testing includes K, Fya, Fyb, Jka, Symptoms Jkb, S, s (k, cellano, testing performed if indicated) to assess maternal or paternal RBC Transfusion reactions or HDFN can occur due to alloimmunization: phenotype status. Antigen Testing, Rh Phenotype 0013019 Intravascular hemolysis: hemoglobinuria, jaundice, shock Extravascular hemolysis: fever and chills Method: Hemagglutination HDFN: fetal hemolytic anemia, hepatosplenomegaly, jaundice, erythroblastosis, Antigen testing for D, C, E, c, and e to assess neurological damage, hydrops fetalis maternal, paternal, or newborn Rh phenotype status Clinical presentation is variable and dependent upon the specific antibody and recipient factors Genotype Testing May help -

Mirepoix LLC on Behalf of Caris Life Sciences, June 22, 2020 No Revisions to These Recommendations
0211U Oncology (pan-tumor), DNA and RNA by next generation sequencing, utilizing formalin-fixed paraffin-embedded tissue, interpretative report for single nucleotide variants, copy number alterations, tumor mutational burden, and microsatellite instability, with therapy association Submitted by Joel de Jesus, Mirepoix LLC on behalf of Caris Life Sciences, June 22, 2020 No revisions to these recommendations 0211U: MI Cancer Seek™ - NGS analysis C. Public Comment Rationale • Recommendation to crosswalk 0211U MI Cancer Seek is a combined whole transcriptome AND whole exome to 0019U (NLA=$3675.00) + 0036U sequencing test that is equivalent to (NLA=$4780.00) performing both: • 0019U: Oncology, RNA, gene expression • 1x multiplier for each by whole transcriptome sequencing, formalin-fixed paraffin-embedded tissue • Recommendation of an NLA equal to or fresh frozen tissue, predictive $8455 for 0211U algorithm reported as potential targets for therapeutic agents • 0036U: Exome (ie, somatic mutations), paired formalin-fixed paraffin-embedded tumor tissue and normal specimen, sequence analyses Submitted by Joel de Jesus, Mirepoix LLC on behalf of Caris Life Sciences, June 22, 2020 No revisions to these recommendations 0211U: MI Cancer Seek™ - NGS analysis C. 0019U* 0211U 0036U** Assay type Whole Whole Whole Whole Transcriptome Transcriptome Exome Exome Sequencing Sequencing Sequencing Sequencing Platform NGS NGS (Illumina) NGS (Illumina) RNASeq RNASeq & DNASeq DNASeq Samples tested FFPE & frozen tumor tissue FFPE tumor tissue FFPE tumor tissue -
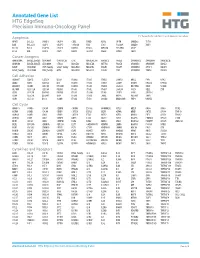
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

Systematic Identification of Cancer Cell Vulnerabilities to Natural Killer Cell
bioRxiv preprint doi: https://doi.org/10.1101/597567; this version posted July 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 2 Systematic identification of cancer cell vulnerabilities to natural killer 3 cell-mediated immune surveillance 4 5 Matthew F. Pech, Linda E. Fong, Jacqueline E. Villalta, Leanne J.G. Chan, Samir Kharbanda, 6 Jonathon J. O’Brien, Fiona E. McAllister, Ari J. Firestone, Calvin H. Jan, Jeff Settleman# 7 8 Calico Life Sciences LLC, 1170 Veterans Boulevard, South San Francisco, CA, United States 9 #correspondence: [email protected] 10 1 bioRxiv preprint doi: https://doi.org/10.1101/597567; this version posted July 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 11 Abstract 12 13 Only a subset of cancer patients respond to T-cell checkpoint inhibitors, highlighting the need for 14 alternative immunotherapeutics. We performed CRISPR-Cas9 screens in a leukemia cell line to 15 identify perturbations that enhance natural killer effector functions. Our screens defined critical 16 components of the tumor-immune synapse and highlighted the importance of cancer cell 17 interferon-g signaling in modulating NK activity. Surprisingly, disrupting the ubiquitin ligase 18 substrate adaptor DCAF15 strongly sensitized cancer cells to NK-mediated clearance. -

Genome-Wide Screen Reveals WNT11, a Non-Canonical WNT Gene, As a Direct Target of ETS Transcription Factor ERG
Oncogene (2011) 30, 2044–2056 & 2011 Macmillan Publishers Limited All rights reserved 0950-9232/11 www.nature.com/onc ORIGINAL ARTICLE Genome-wide screen reveals WNT11, a non-canonical WNT gene, as a direct target of ETS transcription factor ERG LH Mochmann1, J Bock1, J Ortiz-Ta´nchez1, C Schlee1, A Bohne1, K Neumann2, WK Hofmann3, E Thiel1 and CD Baldus1 1Department of Hematology and Oncology, Charite´, Campus Benjamin Franklin, Berlin, Germany; 2Institute for Biometrics and Clinical Epidemiology, Charite´, Campus Mitte, Berlin, Germany and 3Department of Hematology and Oncology, University Hospital Mannheim, Mannheim, Germany E26 transforming sequence-related gene (ERG) is a current therapies in acute leukemia patients with poor transcription factor involved in normal hematopoiesis prognosis characterized by high ERG mRNA expression. and is dysregulated in leukemia. ERG mRNA over- Oncogene (2011) 30, 2044–2056; doi:10.1038/onc.2010.582; expression was associated with poor prognosis in a subset published online 17 January 2011 of patients with T-cell acute lymphoblastic leukemia (T-ALL) and acute myeloid leukemia (AML). Herein, a Keywords: ETS-related gene (ERG); WNT11; acute genome-wide screen of ERG target genes was conducted leukemia; ERG target genes; 6-bromoindirubin-3-oxime by chromatin immunoprecipitation-on-chip (ChIP-chip) in (BIO); morphological transformation Jurkat cells. In this screen, 342 significant annotated genes were derived from this global approach. Notably, ERG-enriched targets included WNT signaling genes: WNT11, WNT2, WNT9A, CCND1 and FZD7. Further- more, chromatin immunoprecipitation (ChIP) of normal Introduction and primary leukemia bone marrow material also confirmed WNT11 as a target of ERG in six of seven The erythroblastosis virus E26 transforming sequence patient samples. -

An Introduction to Blood Groups
1405153490_4_001.qxd 8/16/06 9:21 AM Page 1 An introduction to blood groups CHAPTER 1 What is a blood group? In 1900, Landsteiner showed that people could be divided into three groups (now called A, B, and O) on the basis of whether their red cells clumped when mixed with separated sera from people. A fourth group (AB) was soon found. This is the origin of the term ‘blood group’. A blood group could be defined as, ‘An inherited character of the red cell surface, detected by a specific alloantibody’. Do blood groups have to be present on red cells? This is the usual meaning, though platelet- and neutrophil-specific antigens might also be called blood groups. In this book only red cell surface antigens are considered. Blood groups do not have to be red cell specific, or even blood cell specific, and most are also detected on other cell types. Blood groups do have to be detected by a specific antibody: polymorphisms suspected of being present on the red cell surface, but only detected by other means, such as DNA sequencing, are not blood groups. Furthermore, the antibodies must be alloantibod- ies, implying that some individuals lack the blood group. Blood group antigens may be: • proteins; • glycoproteins, with the antibody recognising primarily the polypeptide backbone; • glycoproteins, with the antibody recognising the carbohydrate moiety; • glycolipids, with the antibody recognising the carbohydrate portion. Blood group polymorphisms may be as fundamental as representing the presence or absence of the whole macromolecule (e.g. RhD), or as minor as a single amino acid change (e.g. -

Characterisation of Weak and Null Phenotypes in the KEL and JK Blood Group Systems
Characterisation of weak and null phenotypes in the KEL and JK blood group systems Sjöberg Wester, Elisabet 2010 Link to publication Citation for published version (APA): Sjöberg Wester, E. (2010). Characterisation of weak and null phenotypes in the KEL and JK blood group systems. Department of Laboratory Medicine, Lund University. Total number of authors: 1 General rights Unless other specific re-use rights are stated the following general rights apply: Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal Read more about Creative commons licenses: https://creativecommons.org/licenses/ Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. LUND UNIVERSITY PO Box 117 221 00 Lund +46 46-222 00 00 Download date: 06. Oct. 2021 Elisabet Sjöberg Wester Elisabet Sjöberg Wester Characterisation of weak and null phenotypes in the KEL and JK blood group systems Characterisation of weak