Long Noncoding Rnas Sustain High Expression of Exogenous Oct4 by Sponging Mirna During Reprogramming
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Filed By: [email protected], Filed Date: 1/7/20 11:04 PM, Submission Status: Approved Page 47 of 123 Barcode:3927422-02 A-351-853 INV - Investigation
Barcode:3927422-02 A-351-853 INV - Investigation - Company Name Address E-mail Phone Website Estrada Municipal - CDR 455, S / N | km 1 Castilian 55 49 3561-3248 and 55- Adami S/A Madeiras Caçador (SC) | Postal Code 89514-899 B [email protected] 49-9184-1887 http://www.adami.com.br/ Rua Distrito Industrial - Quadra 06 - lote 03 - Setor D, Advantage Florestal Ananindeua - PA, 67035-330, Brazil [email protected] 55(91) 3017-5565 https://advantageflorestal.com.br/contact-us/ São Josafat, 1850 Street - Clover - Prudentópolis AFFONSO DITZEL & CIA LTDA Paraná - Brazil - ZIP Code 84400-000 [email protected] 55 42 3446-1440 https://www.affonsoditzel.com/index.php AG Intertrade [email protected] 55 41 3015-5002 http://www.agintertrade.com.br/en/home-2/ General Câmara Street, 243/601 55-51-2217-7344 and Araupel SA 90010-230 - Porto Alegre, RS - Brazil [email protected] 55-51-3254-8900 http://www.araupel.com.br/ Rua Félix da Cunha, 1009 – 8º andar CEP: 90570-001 [email protected] and 55 43 3535-8300 and 55- Braspine Madeiras Ltda. Porto Alegre – RS [email protected] 42-3271-3000 http://www.braspine.com.br/en/home/ R. Mal. Floriano Peixoto, 1811 - 12° andar, Sala 124 - Brazil South Lumber Centro, Guarapuava - PR, 85010-250, Brazil [email protected] 55 42 3622-9185 http://brazilsouthlumber.com.br/?lang=en Curupaitis Street, 701 - Curitiba - Paraná - Brazil - ZIP COMERCIAL EXPORTADORA WK LTDA Code 80.310-180 [email protected] http://wktrading.com.br/ 24 de Outubro Street, -

An Overview of 30 Retail Locations in China Retail Sales in 2013
China City Profiles 2014 An Overview of 30 Retail Locations in China Retail sales in 2013 Y-o-y Growth (%) Beijing 8.7% Shanghai 8.2% Guangzhou 15.2% Chongqing 11.9% Tianjin 14.0% Shenzhen 10.6% Wuhan 13.0% Chengdu 13.1% Suzhou 11.5% Hangzhou 19.9% Nanjing 13.8% Shenyang 13.7% Qingdao 13.3% Changsha 14.1% Wuxi 12.9% Harbin 13.9% Fuzhou 18.7% Ningbo 13.2% Ji'nan 13.4% Zhengzhou 12.9% Xi'an 14.0% Dalian 13.6% Foshan 12.1% Changchun 13.3% Nantong 12.8% Kunming 14.0% Changzhou 13.7% Hefei 14.5% Xiamen 10.5% Zhuhai 13.4% 0 100 200 300 400 500 600 700 800 900 1,000 (RMB billion) Source: CEIC Disposable income in 2013 Y-o-y Growth (%) Shenzhen 9.6% Shanghai 9.1% Guangzhou 10.5% Ningbo 10.1% Xiamen 10.1% Suzhou 9.5% Beijing 10.6% Nanjing 9.8% Hangzhou 4.8% Wuxi 9.4% Foshan 10.0% Changzhou 10.0% Zhuhai 10.3% Ji'nan 9.5% Qingdao 9.6% Changsha 11.1% Xi'an 10.4% Tianjin 10.2% Fuzhou 9.7% Nantong 9.8% Dalian 9.8% Chengdu 10.2% Wuhan 10.2% Shenyang 9.6% Kunming 10.3% Hefei 10.4% Zhengzhou 5.2% Changchun 12.8% Chongqing 9.8% Harbin 12.0% 0 5,000 10,000 15,000 20,000 25,000 30,000 35,000 40,000 45,000 50,000 (RMB per annum) Source: CEIC 2 China City Profiles 2014 China Retail Profiles 2014 The China market presents a compelling opportunity for retailers. -

5 Environmental Baseline
E2646 V1 1. Introduction Public Disclosure Authorized 1.1. Project Background The proposed Harbin-Jiamusi (HaJia Line hereafter) Railway Project is a new 342 km double track railway line starting from the city of Harbin, running through Bing County, Fangzheng County, Yilan County, and ending at the city of Jiasmusi. The Project is located in Heilongjiang Province, and the south of the Songhua River, in the northeast China (See Figure 1-1). The total investment of the Project is RMB 38.66 Billion Yuan, including a World Bank loan of USD 300 million. The construction period is expected to last 4 years, commencing in July 2010. Commissioning of the line is proposed by June 2014. Public Disclosure Authorized HaJia Line, as a Dedicated Passenger Line (DPL) for inter-city communications and an important part of the fast passenger transportation network in northeast of China will extend the Harbin-Dalian dedicated passenger Line to the the northeastern area of Heilongjiang Province, and will be the key line for the transportation system in Heilongjiang Province to go beyond. The project will bring together more closely than before Harbin , Jiamusi and Tongjiang, Shuangyashan, Hegang, Yinchun among which there exists a busy mobility of people potentially demanding high on passenger transportation. The completion of the project will make it possible for the passenger line and cargo train line between Harbin and Jiamusi to be separated, and will extend the the Public Disclosure Authorized line Harbin-Dalian passenger line to the northeast of Heilongjiang Province,It willl also strengthen the skeleton of the railway network of the northeastern part of China and optimize the express passenger transportation network of the northeast. -

General Information
General Information Harbin Harbin, lying on eastern of Songnen plain, the bank of Songhua River, is center of politics, economy and culture and traffic hub in Heilongjiang Province. The special historical course and geographical position make it become an exoticism and beautiful city. It is not only gather the north of the historical and cultural minorities, and the integration of Chinese and foreign cultures. China's famous historical and cultural cities and tourist city, known as the "Jiangcheng", "ice city", "pearl under the Swan," and the "Oriental Moscow", and so on. "Harbin ice and snow Festival" is world-renowned winter tourism. Located at latitude 45 ° 20 '~ 46 ° 20' north, longitude 126 ° 15 '~ 127 ° 30'. The total square of Harbin is 56579 square kilometer, Harbin includes 8 districts and 12 counties, such as Nan Gang district, DaoLi district, Dongli district, XiangFang district, SongBei district, HuLan district, DaoWai district, PingFang district, A Cheng,Shang Zhi, Shuang Cheng county, Bin county, FangZheng county, YiLan county, BaYan county, WuChang county, MuLan county, YanShou county, TongHe county. The population of Harbin is 9,353,200. HARBIN (Ha'erbin) is the capital of Heilongjiang Province and probably the most northernmost location that most tourists visit. A city of distinctive character, Harbin is a good place to visit for its winter atmosphere. Boulevards are lined with European-style buildings and white doves flutter their wings in the skyline of the St. Sofia church. Even though the weather can be very cold, the annual ice lantern festival brings many tourists each year, and the outside influence on this city make it a fascinating place to visit.The nature scene of Harbin is charming and gentle, clearly four seasons .The Sun island, Siberia Tiger Park, Stalin park near Songhua River, SongFeng Mountain, ErLong Mountain, YuQuan hunting ground and YaBuli are famous scenic spot. -

World Bank Document
Certificate No.: Guohuanpingzheng Jiazi No. 1703 Public Disclosure Authorized Environmental Impact Assessment For Harbin Cold Weather Smart Public Transportation System Project (Public Transport Corridors Component) Restructuring Stage Public Disclosure Authorized Entrusted by Public Disclosure Authorized Harbin Transportation Bureau Prepared by Heilongjiang Xingye Environmental Protection Technology Co., Ltd. Revised by HJI Group (USA) Public Disclosure Authorized July 2019 Project Name: Harbin Cold Weather Smart Public Transportation System Project (Public Transport Corridors Component) Project No.: HKYBGS - (2013) 008 Entrusted by: Harbin Transportation Bureau Prepared by: Heilongjiang Xingye Environmental Protection Technology Co., Ltd. Legal Representative: Chi Xiaode EIA Certificate: Guohuanpingzheng Jiazi No. 1703 Task Team Leaders: Sun Baini A17030081000 He Chenyan A17030009 Technical Reviewer: Guan Kezhi A17030023 Prepared by No. of Prepared by Duty Title Registration/Qualification Signature Certificate Sun Baini Report preparation Senior engineer A17030081000 PREFACE Urbanization and motorization have led to constant increase in gasoline consumption in China, and energy will become an important factor affecting China's social and economic development in the future. Over a long time, China's urban public transport system has been underdeveloped, and unable to meet the high-quality daily travelling demand of urban residents. The prominent contradictions between supply and demand in urban transport are shown by the incomplete capacity of urban public transport system, the failure of urban public transport system development in keeping simultaneous with urban development, the single structure of urban public transport system, the lack of advanced transport planning concepts, and the lack of public transport-oriented planning experience for most cities. The project is intended to optimize Harbin's urban public transport system, to improve social and economic development, and to alleviate residents' travel difficulties in cold weather. -

ANNUAL REPORT 2013 Vision the Group Aspires to Be a Leading Chinese Property Developer with a Renowned Brand Backed by Cultural Substance
ANNUAL REPORT 2013 Vision The Group aspires to be a leading Chinese property developer with a renowned brand backed by cultural substance. Mission The Group is driven by a corporate spirit and fine tradition that attaches importance to dedication, honesty and integrity. Its development strategy advocates professionalism, market-orientation and internationalism. It also strives to enhance the architectural quality and commercial value of the properties by instilling cultural substance into its property projects. Ultimately, it aims to build a pleasant living environment for its clients and create satisfactory returns to its shareholders. CONTENTS 2 Corporate Information 6 Chairman’s Statement 14 Management Discussion and Analysis 46 Corporate Governance Report 58 Directors’ Profile 60 Directors’ Report 65 Independent Auditor’s Report 67 Consolidated Income Statement 68 Consolidated Statement of Comprehensive Income 69 Consolidated Statement of Financial Position 71 Statement of Financial Position 72 Consolidated Statement of Changes in Equity 73 Consolidated Statement of Cash Flows 76 Notes to the Consolidated Financial Statements 182 Financial Summary 183 Summary of Properties Held for Investment Purposes 187 Summary of Properties Held for Development 194 Summary of Properties Held for Sale 2 CORPORATE INFORMATION Board of Directors Legal Advisor Ashurst Hong Kong Executive directors CHEN Hong Sheng WANG Xu Auditor XUE Ming (Chairman and Managing Director) PKF ZHANG Wan Shun YE Li Wen Principal Bankers Non-executive director China CITIC Bank International Limited IP Chun Chung, Robert Malayan Banking Berhad Agricultural Bank of China Limited Bank of China Limited Independent non-executive directors China Construction Bank Corporation YAO Kang, J . P. (Resigned on 15th May, 2013) Industrial and Commercial Bank of China Limited CHOY Shu Kwan Bank of Communications Co., Ltd. -
Chinese Mainland
Address List of Special Warehousing Service Note: The address marked in red are newly added address. (Effective date:October 1, 2021) Province / Directly- controlled City District/county Town, Sub-district and House Number Municipality / Autonomous Region/SAR B4-25, Gate 1, ProLogis Logistics Park, No.1 Tiedi Road, Anhui Province Hefei Shushan District High-tech Zone No.18 Tianzhushan Road, Longshan Sub-district, Wuhu Anhui Province Wuhu Jiujiang District Economic and Technological Development Zone Anhui Province Chuzhou Langya District Longji Leye Photovoltaic Co., Ltd., No.19 Huai'an Road 3/F, No.8 Building, South Area, Lixiang Innovation Park, Anhui Province Chuzhou Nanqiao District Chuzhou, 018 Township Road Anhui Province Chuzhou Nanqiao District No.19 Huai'an Road Yuanrong New Material Holding Co., Ltd., 50 Meters Anhui Province Hefei Shushan District Westward of Bridge of Intersection of Changning Avenue and Ningxi Road Anhui Province Hefei Yaohai District No.88 Dayu Road Anhui Province Hefei Yaohai District No.2177 Dongfang Avenue Beijing BOE Vision-Electronic Technology Co., Ltd., No. Anhui Province Hefei Yaohai District 2177 Dongfang Avenue Anhui Province Hefei Yaohai District No.668 Longzihu Road Anhui Province Hefei Yaohai District No. 668 Longzihu Road Anhui Province Hefei Yaohai District No.2177 Tongling North Road Anhui Province Hefei Yaohai District No.3166 Tongling North Road Anhui Province Hefei Yaohai District No.8 Xiangwang Road Anhui Province Wuhu Jiujiang District No. 8 Anshan Road Anhui Province Wuhu Jiujiang District -

Study on the Preparation and Conjugation Mechanism of the Phosvitin-Gallic Acid Complex with an Antioxidant and Emulsifying Capability
polymers Article Study on the Preparation and Conjugation Mechanism of the Phosvitin-Gallic Acid Complex with an Antioxidant and Emulsifying Capability Bin Jiang 1 , Xiaojing Wang 1, Linlin Wang 1, Shuang Wu 2, Dongmei Li 1, Chunhong Liu 1 and Zhibiao Feng 1,* 1 Department of Applied Chemistry, Northeast Agricultural University, NO.600 Changjiang Road Xiangfang District, Harbin 150030, China 2 Heilongjiang Eco-meteorology Center, NO.71 Diantan Road Xiangfang District, Harbin 150030, China * Correspondence: [email protected]; Tel.: +86-4515-519-0222 Received: 13 August 2019; Accepted: 5 September 2019; Published: 7 September 2019 Abstract: To develop a novel emulsifier with an antioxidant capacity, a phosvitin-gallic acid (Pv–GA) complex was prepared via a free-radical method. This emulsifier characterizes some key technologies. Changes in the molecular weight of the Pv–GA complex were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) and the matrix-assisted laser desorption/ionization time of light mass spectrometry (MALDI-TOF-MS). Fourier transform infrared spectroscopy (FTIR) indicated that C=O, C–N and N–H groups were also likely to be involved in the formation of the complex. A redshift was obtained in the fluorescence spectrogram, thereby proving that the covalent combination of Pv and GA was a free radical-forming complex. The results indicated that Pv and GA were successfully conjugated. Meanwhile, the secondary structure of Pv showed significant changes after conjugation with GA. The antioxidant activity and emulsifying properties of the Pv–GA complex were studied. The antioxidant activity of the Pv–GA complex proved to be much higher than that of the Pv, via assays of the scavenging activities of 2,2-Diphenyl-1-picrylhydrazyl (DPPH) and 2,2’-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) radicals and because of their ability to reduce power. -

Chinese Fuel Saving Stoves: a Compendium
Field Document No.40 REGIONAL WOOD ENERGY DEVELOPMENT PROGRAMME IN ASIA GCP/RAS/154/NET CHINESE FUEL SAVING STOVES: A COMPENDIUM FOOD AND AGRICULTURE ORGANIZATION OF THE UNITED NATIONS Bangkok, July 1993 This publication is printed by the FAO Regional Wood Energy Development Programme in Asia, Bangkok, Thailand The designations employed and the presentation of material in this publication do not imply the expression of any opinion whatsoever on the part of the Food and Agriculture Organiza- tion of the United nations concerning the legal status of any country, territory, city or area or of its authorities, or concerning the delimitations of its frontiers or boundaries. The opinions expressed in this publication are those of the author(s) alone and do not imply any opinion on the part of the FAO. For copies write to: Regional Wood Energy Development Programme in Asia Tel: 66-2-280 2760 c/o FAO Regional Offcie for Asia and the Pacific Fax: 66-2-280 0760 Maliwan Mansion, Phra Atit Road, E-mail: [email protected] Bangkok, Thailand Internet: http://www.rwedp.org FOREWORD Most countries in Asia are giving an increasingly higher priority to energy conservation, both in the industrial/commercial and domestic sectors. As cooking in developing countries constitutes a large part of the total energy consumption in the domestic sector, conservation approaches have concentrated on the development and introduction of improved cooking stoves. From its very beginning in 1985 the Regional Wood Energy Development Programme in Asia (RWEDP) has supported or initiated activities in the field of improved cookstoves development, with particular emphasis on information sharing and transfer of knowledge. -
HARBIN the “Ice City” in Summer, Hot and Cold
HARBIN The “Ice City” in summer, hot and cold Location of Harbin Harbin is the birthplace of the Jurchen Jin Dynasty and is currently one of the political, economic and cultural centres of Northeast China. It is also a hot tourist city in the northeast and an international city famous with ice and snow culture, known as the “Ice City” and “Moscow of the East”. Harbin has four distinct seasons, but due to its high latitude, winter is long and summer is short, with average temperatures of 19 to 23°C in summer, making it a famous summer destination. While visiting the cool local attractions, visitors can also experience the lively summer holiday atmosphere, enjoy the cold and heat of the Ice City. What’s hot Harbin Polarpark Harbin Polarpark is one of the four major attractions of the Harbin International Ice and Snow Sculpture Festival. It is the first amusement park to provide polar themed entertainment in the world, with several classic original polar themed shows and a wide range of North and South Pole animals. In addition to the world’s only Beluga Show, the Penguin Scene Play, Ray Fish Show, Walrus and Sea Lion Show, there are many other dazzling music performances as well. One can also visit the newly opened Dinosaur Hall and meet dinosaurs from the Jurassic period from late July 2021. 3 Sun Avenue, Songbei District, Harbin City, Heilongjiang Province, China (near the main entrance of Sun Island Park) Take bus Route 42, tour bus Line 1, 2, or 3 and get off at Taiyangdao Stop, then walk about 100 metres. -
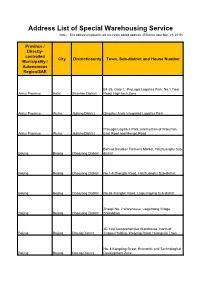
Address List of Special Warehousing Service Note: the Address Marked in Red Are Newly Added Address
Address List of Special Warehousing Service Note: The address marked in red are newly added address. (Effective date:Mar. 28, 2019) Province / Directly- controlled City District/county Town, Sub-district and House Number Municipality / Autonomous Region/SAR B4-25, Gate 1, ProLogis Logistics Park, No.1 Tiedi Anhui Province Hefei Shushan District Road, High-tech Zone Anhui Province Wuhu Jiujiang District Qingshui Ande Integrated Logistics Park ProLogis Logistics Park, Intersection of Wanchun Anhui Province Wuhu Jiujiang District East Road and Mengxi Road Behind Daludian Farmer's Market, Heizhuanghu Sub- Beijing Beijing Chaoyang District district Beijing Beijing Chaoyang District No.1-9 Zhangtai Road, Heizhuanghu Sub-district Beijing Beijing Chaoyang District No.66 Xiangbin Road, Laiguangying Sub-district Zhaopi No. 2 Warehouse, Laojuntang Village, Beijing Beijing Chaoyang District Shibalidian JD Yayi Comprehensive Warehouse, North of Beijing Beijing Daxing District Jingwei Holding, Weiyong Road, Huangcun Town No. 8 Kangding Street, Economic and Technological Beijing Beijing Daxing District Development Zone Beijing Beijing Daxing District No. 70 Weiyong Road Beijing Beijing Daxing District CEIEC, Xinghai 1st Street No. 11 Jinxiu Street, Yizhuang Economic and Technological Development Zone/No. 18 Yuncheng Street, Yizhuang Economic and Technological Beijing Beijing Daxing District Development Zone Beijing Beijing Daxing District No. 8 Kangding Street, Yizhuang Beijing Beijing Fengtai District No.102 Yangshuzhuang No. 3 Area F2D, Jinxiudadi -

Because Tomorrow Matters
BECAUSE TOMORROW MATTERS ANNUAL REPORT 20 20 Underpinning the breadth of CapitaLand China Trust's activities is our continuous dialogue with our stakeholders which helps to shape our business as we aim to build sustainable communities. We place significance on our conversations and share our story with care and consideration for all involved. This annual report is part of that process and the motif we have chosen for this year’s report reflects our focus on maintaining communication with our stakeholders with transparency and clarity. WE WILL CONTINUE TO SHARPEN OUR STRATEGIES, EMBRACE NEW OPPORTUNITIES AND POSITION OURSELVES AS THE PROXY FOR GROWTH IN CHINA’S FUTURE ECONOMY. TAN TZE WOOI CEO CAPITALAND CHINA TRUST MANAGEMENT LIMITED BECAUSE TOMORROW MATTERS BECAUSE CHANGE CREATES OPPORTUNITY GLOBAL EVENTS THIS YEAR ACCELERATED THE NEED FOR BUSINESSES TO DEVELOP INNOVATIVE WAYS TO STAY AHEAD. AT CLCT, WE EMBRACED NEW MEANS OF CUSTOMER ENGAGEMENT, STRENGTHENED CONNECTIONS THROUGH OMNICHANNEL MARKETING INITIATIVES, AND LEVERAGED ON DIGITALISATION WITH OUR ESTABLISHED CAPITASTAR PROGRAMME. STAYING NIMBLE TO CAPTURE EVOLVING RETAIL TRENDS HELPS TO FUTURE PROOF OUR BUSINESS AND ENHANCE CUSTOMERS’ STICKINESS TO OUR MALLS. BECAUSE EXPANSION OPENS POSSIBILITIES WE EXPANDED OUR INVESTMENT MANDATE TO COVER RETAIL, OFFICE AND INDUSTRIAL ASSETS, INCLUDING BUSINESS PARKS, LOGISTICS FACILITIES, DATA CENTRES, AND INTEGRATED DEVELOPMENTS. CLCT IS THE DEDICATED SINGAPORE-LISTED REIT (S-REIT) FOR CAPITALAND GROUP’S NON-LODGING CHINA BUSINESSES, WITH ACQUISITION PIPELINE ACCESS TO CAPITALAND’S CHINA ASSETS. WE CAN CAPTURE OPPORTUNITIES FROM BOTH OUR SPONSOR AND THIRD-PARTY VENDORS TO RIDE THE WAVE OF CHINA’S CONSUMPTION-DRIVEN, HIGHER-VALUED, SERVICE-LED ECONOMY.