Discrimination and Genetic Diversity of Cephalotaxus Accessions Using AFLP Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cephalotaxus
Reprinted from the Winter 1970 issue of t'he THE AMERICA HORTICULTURAL \t{AGAZIl\'E Copyright 1970 by The American Horticultural Society, Inc. Cephalotaxus Source of Harringtonine, A Promising New Anti..Cancer Alkaloid ROBERT E. PERDUE, JR.,l LLOYD A. SPETZMAN,l and RICHARD G. POWELL2 The plumyews (Cephalotaxus) are yew-like evergreen trees and shrubs. The genus includes seven species native to southeastern Asia from Japan and Korea to Taiwan and Hainan, and west through China to northeastern India. Two species are in cultivation in the United States, C. harringtoniaJ (Fig. 1 & 2) of which there are several varieties (one often listed as C. drupacea) , and C. fortunii (Fig. 3). The cultivars are shrubs up to about 20 feet in height; most have broad crowns. The linear and pointed leaves are spirally arranged or in two opposite ranks. The upper sur Fig. 1. Japanese plumyew (Cephalo face is dark shiny green with a conspicu taxus harringtonia var. drupacea), an ous mid-rib; the lower surface has a evergreen shrub about 6 ft. high, at broad silvery band on either side of the the USDA Plant Introduction Station, mid-rib. These bands are made up of Glenn Dale, Maryland. This photo conspicuous white stomata arranged in graph was made in 1955. The plant is numerous distinct lines. Leaf length is now about 7 ft. high, but the lower variable, from about one inch in varie branches have been severely pruned ties of C. harringtonia to three or four to provide material for chemical re inches in C. fortunii. The leaves are search. -

Torreya Taxifolia
photograph © Abraham Rammeloo Torreya taxifolia produces seeds in 40 Kalmthout Arboretum ABRAHAM RAMMELOO, Curator of the Kalmthout Arboretum, writes about this rare conifer that recently produced seed for the first time. Torreya is a genus of conifers that comprises four to six species that are native to North America and Asia. It is closely related to Taxus and Cephalotaxus and is easily confused with the latter. However, it is relatively easy to distinguish them apart by their leaves. Torreya has needles with, on the underside, two small edges with stomas giving it a green appearance; Cephalotaxus has different rows of stomas, and for this reason the underside is more of a white colour. It is very rare to find Torreya taxifolia in the wild; it is native to a small area in Florida and Georgia. It grows in steep limestone cliffs along the Apalachicola River. These trees come from a warm and humid climate where the temperature in winter occasionally falls below freezing. They grow mainly on north-facing slopes between Fagus grandifolia, Liriodendron tulipifera, Acer barbatum, Liquidambar styraciflua and Quercus alba. They can grow up to 15 to 20 m high. The needles are sharp and pointed and grow in a whorled pattern along the branches. They are 25 to 35 mm long and stay on the tree for three to four years. If you crush them, they give off a strong, sharp odour. The health and reproduction of the adult population of this species suffered INTERNATIONAL DENDROLOGY SOCIETY TREES Opposite Torreya taxifolia ‘Argentea’ growing at Kalmthout Arboretum in Belgium. -
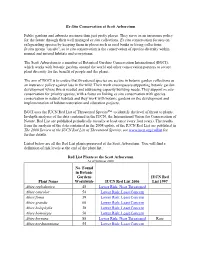
IUCN Red List of Threatened Species™ to Identify the Level of Threat to Plants
Ex-Situ Conservation at Scott Arboretum Public gardens and arboreta are more than just pretty places. They serve as an insurance policy for the future through their well managed ex situ collections. Ex situ conservation focuses on safeguarding species by keeping them in places such as seed banks or living collections. In situ means "on site", so in situ conservation is the conservation of species diversity within normal and natural habitats and ecosystems. The Scott Arboretum is a member of Botanical Gardens Conservation International (BGCI), which works with botanic gardens around the world and other conservation partners to secure plant diversity for the benefit of people and the planet. The aim of BGCI is to ensure that threatened species are secure in botanic garden collections as an insurance policy against loss in the wild. Their work encompasses supporting botanic garden development where this is needed and addressing capacity building needs. They support ex situ conservation for priority species, with a focus on linking ex situ conservation with species conservation in natural habitats and they work with botanic gardens on the development and implementation of habitat restoration and education projects. BGCI uses the IUCN Red List of Threatened Species™ to identify the level of threat to plants. In-depth analyses of the data contained in the IUCN, the International Union for Conservation of Nature, Red List are published periodically (usually at least once every four years). The results from the analysis of the data contained in the 2008 update of the IUCN Red List are published in The 2008 Review of the IUCN Red List of Threatened Species; see www.iucn.org/redlist for further details. -

Complete Chloroplast Genome of Fokienia Hodginsii (Dunn) Henry Et Thomas: Insights Into Repeat Regions Variation and Phylogenetic Relationships in Cupressophyta
Article Complete Chloroplast Genome of Fokienia hodginsii (Dunn) Henry et Thomas: Insights into Repeat Regions Variation and Phylogenetic Relationships in Cupressophyta Mingyue Zang 1 , Qian Su 1, Yuhao Weng 1, Lu Lu 1, Xueyan Zheng 2, Daiquan Ye 2, Renhua Zheng 3, Tielong Cheng 1,4, Jisen Shi 1 and Jinhui Chen 1,4,* 1 Key Laboratory of Forest Genetics & Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, Nanjing 210037, China; [email protected] (M.Z.); [email protected] (Q.S.); [email protected] (Y.W.); [email protected] (L.L.); [email protected] (T.C.); [email protected] (J.S.) 2 National Germplasm Bank of Chinese fir at Fujian Yangkou Forest Farm, Shunchang 353211, China; [email protected] (X.Z.); [email protected] (D.Y.) 3 The Key Laboratory of Timber Forest Breeding and Cultivation for Mountainous Areas in Southern China, State Forestry Administration Engineering Research Center of Chinese Fir, Fujian Academy of Forestry, Fuzhou 350012, China; [email protected] 4 College of Biology and the Environment, Nanjing Forestry University, Nanjing 210037, China * Correspondence: [email protected]; Tel./Fax: +86-25-85428948 Received: 24 May 2019; Accepted: 21 June 2019; Published: 26 June 2019 Abstract: Fokienia hodginsii (Dunn) Henry et Thomas is a relic gymnosperm with broad application value. It is a fit candidate when choosing species for the construction of artificial forests. We determined the complete chloroplast genome sequence of F. hodginsii, which is 129,534 bp in length and encodes 83 protein genes, 33 transfer RNA (tRNA) genes, as well as four ribosomal RNA genes. -

Environmental Impact Analysis in This Report
Environmental Impacts Assessment Report on Project Construction Project name: European Investment Bank Loan Hunan Camellia Oil Development Project Construction entity (Seal): Foreign Fund Project Administration Office of Forestry Department of Hunan Province Date of preparation: July 1st, 2012 Printed by State Environmental Protection Administration of China Notes for Preparation of Environmental Impacts Assessment Report on Project Construction An Environmental Impacts Assessment (EIA) Report shall be prepared by an entity qualified for conducting the work of environmental impacts assessment. 1. Project title shall refer to the name applied by the project at the time when it is established and approved, which shall in no case exceed 30 characters (and every two English semantic shall be deemed as one Chinese character) 2. Place of Construction shall refer to the detailed address of project location, and where a highway or railway is involved, names of start station and end station shall be provided. 3. Industry category shall be stated according to the Chinese national standards. 4. Total Investment Volume shall refer to the investment volume in total of the project. 5. Principal Targets for Environment Protection shall refer to centralized residential quarters, schools, hospitals, protected culture relics, scenery areas, water sources and ecological sensitive areas within certain radius of the project area, for which the objective, nature, size and distance from project boundary shall be set out as practical as possible. 6. Conclusion and suggestions shall include analysis results for clean production, up-to-standard discharge and total volume control of the project; a determination on effectiveness of pollution control measures; an explanation on environmental impacts by the project, and a clear-cut conclusion on feasibility of the construction project. -

Characterization of 15 Polymorphic Microsatellite Loci for Cephalotaxus Oliveri (Cephalotaxaceae), a Conifer of Medicinal Importance
Int. J. Mol. Sci. 2012, 13, 11165-11172; doi:10.3390/ijms130911165 OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Short Note Characterization of 15 Polymorphic Microsatellite Loci for Cephalotaxus oliveri (Cephalotaxaceae), a Conifer of Medicinal Importance Yingchun Miao 1,2, Xuedong Lang 2, Shuaifeng Li 2, Jianrong Su 2,* and Yuehua Wang 1,* 1 Department of Botany, School of Life Sciences, Yunnan University, Kunming 650091, China; E-Mail: [email protected] 2 Research Institute of Resource Insects, Chinese Academy of Forest (CAF), Kunming 650224, China; E-Mails: [email protected] (X.L.); [email protected] (S.L.) * Authors to whom correspondence should be addressed; E-Mails: [email protected] (J.S.); [email protected] (Y.W.); Tel.: +86-871-3860017; Fax: +86-871-3860017. Received: 7 August 2012; in revised form: 28 August 2012 / Accepted: 2 September 2012 / Published: 7 September 2012 Abstract: Cephalotaxus oliveri is a scarce medicinal conifer endemic to the south central region of China and Vietnam. A small fragmented population presently exists due to anthropogenic disturbance. C. oliveri has been used for its alkaloids harringtonine and homoharringtonine, which are effective against leucocythemia and lymphadenosarcoma. Monoecious plants have been detected in nature, although they were understood to be dioecious. In order to study the mating system, population genetics and the genetic effects of habitat fragmentation on C. oliveri, 15 polymorphic and 12 monomorphic microsatellite loci were developed for C. oliveri by using the Fast Isolation by AFLP of Sequences Containing repeats (FIASCO) protocol. The polymorphisms were assessed in 96 individuals from three natural populations (32 individuals per population). -

A Comparative Study of the Primary Vascular System Of
ArneI'. J. Bot. 5.5(4): 447-457. 1968. A COMPARATIVE STUDY OF THE PRIMARY VASCULAR SYSTElVI OF CONIFERS. 1. GENERA WITH HELICAL PHYLLOTAXISl KADAMBARI K. N AMBOODIRI2 AND CHARLES B. BECK Department of Botany, University of Michigan, Ann Arbor ABSTRACT The primary vascular system of 23 species belonging to 18 genera of conifers with helical phyllotaxis has been investigated with the intent of determining the architecture .f the system. Special attention has been given to nodal and subnodal relations of the vascular bundles. The vascular system seems to be composed solely of relatively discrete sympodia, that is, axial vascu lar bundles from which leaf traces branch unilaterally. Although the discreteness of the syrn podia is not immediately apparent because of their undulation and lateral contacts with neigh boring ones, close examination, including a statistical analysis of the tangential contacts, seems to reveal that each sympodium maintains its identity throughout. Although two traces may be apparent at nodal levels, the trace supply to a leaf originates, in all species, as a single bundle. An analysis is made of the relationship between the vasculature and the phyllotaxis. It is ob served that the direction of trace divergence can be accurately predicted when the direction of the ontogenetic spiral, the angle of divergence of leaf traces, and the number of syrnpodia are known. THE ORIGIN and evolution of gymnosperms that of the ferns by reduction (Jeffrey, 1902, are significant problems that deserve increased 1917). Consequently, he considered the leaf gap attention. There have been few modern compara of seed plants to be homologous with that of tive studies of extant gymnosperms, and most the ferns. -

Number 3, Spring 1998 Director’S Letter
Planning and planting for a better world Friends of the JC Raulston Arboretum Newsletter Number 3, Spring 1998 Director’s Letter Spring greetings from the JC Raulston Arboretum! This garden- ing season is in full swing, and the Arboretum is the place to be. Emergence is the word! Flowers and foliage are emerging every- where. We had a magnificent late winter and early spring. The Cornus mas ‘Spring Glow’ located in the paradise garden was exquisite this year. The bright yellow flowers are bright and persistent, and the Students from a Wake Tech Community College Photography Class find exfoliating bark and attractive habit plenty to photograph on a February day in the Arboretum. make it a winner. It’s no wonder that JC was so excited about this done soon. Make sure you check of themselves than is expected to seedling selection from the field out many of the special gardens in keep things moving forward. I, for nursery. We are looking to propa- the Arboretum. Our volunteer one, am thankful for each and every gate numerous plants this spring in curators are busy planting and one of them. hopes of getting it into the trade. preparing those gardens for The magnolias were looking another season. Many thanks to all Lastly, when you visit the garden I fantastic until we had three days in our volunteers who work so very would challenge you to find the a row of temperatures in the low hard in the garden. It shows! Euscaphis japonicus. We had a twenties. There was plenty of Another reminder — from April to beautiful seven-foot specimen tree damage to open flowers, but the October, on Sunday’s at 2:00 p.m. -

Wa Shan – Emei Shan, a Further Comparison
photograph © Zhang Lin A rare view of Wa Shan almost minus its shroud of mist, viewed from the Abies fabri forested slopes of Emei Shan. At its far left the mist-filled Dadu River gorge drops to 500-600m. To its right the 3048m high peak of Mao Kou Shan climbed by Ernest Wilson on 3 July 1903. “As seen from the top of Mount Omei, it resembles a huge Noah’s Ark, broadside on, perched high up amongst the clouds” (Wilson 1913, describing Wa Shan floating in the proverbial ‘sea of clouds’). Wa Shan – Emei Shan, a further comparison CHRIS CALLAGHAN of the Australian Bicentennial Arboretum 72 updates his woody plants comparison of Wa Shan and its sister mountain, World Heritage-listed Emei Shan, finding Wa Shan to be deserving of recognition as one of the planet’s top hotspots for biological diversity. The founding fathers of modern day botany in China all trained at western institutions in Europe and America during the early decades of last century. In particular, a number of these eminent Chinese botanists, Qian Songshu (Prof. S. S. Chien), Hu Xiansu (Dr H. H. Hu of Metasequoia fame), Chen Huanyong (Prof. W. Y. Chun, lead author of Cathaya argyrophylla), Zhong Xinxuan (Prof. H. H. Chung) and Prof. Yung Chen, undertook their training at various institutions at Harvard University between 1916 and 1926 before returning home to estab- lish the initial Chinese botanical research institutions, initiate botanical exploration and create the earliest botanical gardens of China (Li 1944). It is not too much to expect that at least some of them would have had personal encounters with Ernest ‘Chinese’ Wilson who was stationed at the Arnold Arboretum of Harvard between 1910 and 1930 for the final 20 years of his life. -

Gymnosperms on the EDGE Félix Forest1, Justin Moat 1,2, Elisabeth Baloch1, Neil A
www.nature.com/scientificreports OPEN Gymnosperms on the EDGE Félix Forest1, Justin Moat 1,2, Elisabeth Baloch1, Neil A. Brummitt3, Steve P. Bachman 1,2, Stef Ickert-Bond 4, Peter M. Hollingsworth5, Aaron Liston6, Damon P. Little7, Sarah Mathews8,9, Hardeep Rai10, Catarina Rydin11, Dennis W. Stevenson7, Philip Thomas5 & Sven Buerki3,12 Driven by limited resources and a sense of urgency, the prioritization of species for conservation has Received: 12 May 2017 been a persistent concern in conservation science. Gymnosperms (comprising ginkgo, conifers, cycads, and gnetophytes) are one of the most threatened groups of living organisms, with 40% of the species Accepted: 28 March 2018 at high risk of extinction, about twice as many as the most recent estimates for all plants (i.e. 21.4%). Published: xx xx xxxx This high proportion of species facing extinction highlights the urgent action required to secure their future through an objective prioritization approach. The Evolutionary Distinct and Globally Endangered (EDGE) method rapidly ranks species based on their evolutionary distinctiveness and the extinction risks they face. EDGE is applied to gymnosperms using a phylogenetic tree comprising DNA sequence data for 85% of gymnosperm species (923 out of 1090 species), to which the 167 missing species were added, and IUCN Red List assessments available for 92% of species. The efect of diferent extinction probability transformations and the handling of IUCN data defcient species on the resulting rankings is investigated. Although top entries in our ranking comprise species that were expected to score well (e.g. Wollemia nobilis, Ginkgo biloba), many were unexpected (e.g. -

Mitteilungen Der Deutschen Dendrologischen Gesellschaft
: l8 Mitteilungen der Deutschen Dendrologischen Gesellschaft. 1898. Ein leichter Schutz des Erdreichs durch Nadelholzreisig, besonders der etwas empfindlichen Rhododendron ist Vorteil. Ein Schutz gegen strenge Insolation in den angegebenen Zeiten, ist bei sämtlichen immergrünen Gehölzen von grofsem Nutzen. Herr Brei/schwerdt -MdLinz (bis vergangenes Jahr in Donaueschingen in Stellung) bestätigt des Vorredners Mitteilung betreffs der Rhododendron in Donaueschingen und teilt mit, dafs auch die Neupflanzungen dort vortrefflich gedeihen. Nunmehr berichtet Garteninspektor jS'fz/y/^«?;'- Poppelsdorf- Bonn über „Neues und Interessantes über Coniferen" unter Vorlage seltener Coniferen- Zapfen. Neues und Interessantes über Coniferen. Von Ij. Beissner, Königl, Garteninspektor in Poppelsdorf bei Bonn. Seit dem letzten über Coniferen erstatteten Berichte ist wieder manches Be- merkenswerte zu verzeichnen. Zuerst nenne ich eine wohl wenig bekannte und verbreitete Form von Biota nämlich Biota orientalis sphaeroidea glauca, welche ich von Herrn Simon Louis Freres in Plantieres bei Metz erhielt. Es ist eine Kugelform von bläulicher Färbung und bietet dadurch besonders Interesse, dafs sie alle Übergänge in der Bezweigung trägt von der Jugendform decussata zu meldensis bis zur nor- malen orientalis. Solche Exemplare veranschaulichen besonders deutlich die ver- schiedenen Entwickelungsstadien ein und derselben Pflanzenart, welche jede für sich durch Stecklinge fortgepflanzt, für den Nichtkenner als ganz abweichende Individuen, immer noch zu Schwierigkeiten -

Arboretum News Armstrong News & Featured Publications
Georgia Southern University Digital Commons@Georgia Southern Arboretum News Armstrong News & Featured Publications Arboretum News Number 5, Summer 2006 Armstrong State University Follow this and additional works at: https://digitalcommons.georgiasouthern.edu/armstrong-arbor- news Recommended Citation Armstrong State University, "Arboretum News" (2006). Arboretum News. 5. https://digitalcommons.georgiasouthern.edu/armstrong-arbor-news/5 This newsletter is brought to you for free and open access by the Armstrong News & Featured Publications at Digital Commons@Georgia Southern. It has been accepted for inclusion in Arboretum News by an authorized administrator of Digital Commons@Georgia Southern. For more information, please contact [email protected]. Arboretum News A Newsletter of the Armstrong Atlantic State University Arboretum Issue 5 Summer 2006 Watch Your Step in the Primitive Garden Arboretum News Arboretum News, published by the Grounds Department Plants from the Past of Armstrong Atlantic State University, is distributed to Living Relatives of Ancient faculty, staff, students, and friends of the Arboretum. The Arboretum Plants in the Primitive Garden encompasses Armstrong’s 268- acre campus and displays a wide By Philip Schretter variety of shrubs and other woody plants. Developed areas of campus he Primitive Garden, contain native and introduced Tlocated next to Jenkins species of trees and shrubs, the Hall on the Armstrong majority of which are labeled. Atlantic State University Natural areas of campus contain campus, allows you to take plants typical in Georgia’s coastal a walk through time by broadleaf evergreen forests such as displaying living relatives of live oak, southern magnolia, red ancient plants. The following bay, horse sugar, and sparkleberry.