Polyclad Phylogeny Persists to Be Problematic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

(Platyhelminthes, Polycladida, Cotylea) from the Persian Gulf, Iran
A peer-reviewed open-access journal ZooKeys 31: 39–51 (2009)First record of the family Pseudocerotidae the Persian Gulf, Iran 39 doi: 10.3897/zookeys.31.136 RESEARCH ARTICLE www.pensoftonline.net/zookeys Launched to accelerate biodiversity research First record of the family Pseudocerotidae (Platyhelminthes, Polycladida, Cotylea) from the Persian Gulf, Iran Zahra Khalili1, Hassan Rahimian2, Jamile Pazooki1 1 University of Shahid Beheshti, G.C, Tehran, Iran 2 University of Tehran, Tehran, Iran Corresponding authors: Hassan Rahimian ([email protected]), Zahra Khalili ([email protected]) Academic editor: E. Neubert, Z. Amr | Received 7 March 2009 | Accepted 14 August 2009 | Published 28 December 2009 Citation: Khalili Z, Rahimian H, Pazooki J (2009) First record of the family Pseudocerotidae (Platyhelminthes, Po- lycladida, Cotylea) from the Persian Gulf, Iran. In: Neubert, E, Amr, Z, Taiti, S, Gümüs, B (Eds) Animal Biodiversity in the Middle East. Proceedings of the First Middle Eastern Biodiversity Congress, Aqaba, Jordan, 20–23 October 2008. ZooKeys 31: 39–51. doi: 10.3897/zookeys.31.136 Abstract In this paper, two species of cotylean Platyhelminthes are recorded for the fi rst time from Qeshm Island, Persian Gulf, Iran. Pictures are taken from living specimens to illustrate shape and colour, and stained sec- tions and drawings are used to describe shape and organisation of some organs. Morphological characters of Persian Gulf specimens of Tytthosoceros lizardensis Newman and Cannon 1996 are compared to those of the type specimens of this species. Keywords Platyhelminthes, new records, Qeshm Island, Persian Gulf, Iran Introduction Most polyclad fl atworms inhabit coral reefs in tropical and subtropical waters, and are espe- cially species-rich throughout the Indo-Pacifi c. -

Natural Products in Polyclad Flatworms
marine drugs Review Natural Products in Polyclad Flatworms Justin M. McNab 1 , Jorge Rodríguez 1, Peter Karuso 2,* and Jane E. Williamson 1,* 1 Department of Biological Sciences, Macquarie University, Sydney, NSW 2109, Australia; [email protected] (J.M.M.); [email protected] (J.R.) 2 Department of Molecular Sciences, Macquarie University, Sydney, NSW 2109, Australia * Correspondence: [email protected] (P.K.); [email protected] (J.E.W.) Abstract: Marine invertebrates are promising sources of novel bioactive secondary metabolites, and organisms like sponges, ascidians and nudibranchs are characterised by possessing potent defensive chemicals. Animals that possess chemical defences often advertise this fact with aposematic colouration that potential predators learn to avoid. One seemingly defenceless group that can present bright colouration patterns are flatworms of the order Polycladida. Although members of this group have typically been overlooked due to their solitary and benthic nature, recent studies have isolated the neurotoxin tetrodotoxin from these mesopredators. This review considers the potential of polyclads as potential sources of natural products and reviews what is known of the activity of the molecules found in these animals. Considering the ecology and diversity of polyclads, only a small number of species from both suborders of Polycladida, Acotylea and Cotylea have been investigated for natural products. As such, confirming assumptions as to which species are in any sense toxic or if the compounds they use are biosynthesised, accumulated from food or the product of symbiotic bacteria is difficult. However, further research into the group is suggested as these animals often display aposematic colouration and are known to prey on invertebrates rich in bioactive secondary metabolites. -

Platyhelminthes) at the Queensland Museum B.M
VOLUME 53 ME M OIRS OF THE QUEENSLAND MUSEU M BRIS B ANE 30 NOVE mb ER 2007 © Queensland Museum PO Box 3300, South Brisbane 4101, Australia Phone 06 7 3840 7555 Fax 06 7 3846 1226 Email [email protected] Website www.qm.qld.gov.au National Library of Australia card number ISSN 0079-8835 Volume 53 is complete in one part. NOTE Papers published in this volume and in all previous volumes of the Memoirs of the Queensland Museum may be reproduced for scientific research, individual study or other educational purposes. Properly acknowledged quotations may be made but queries regarding the republication of any papers should be addressed to the Editor in Chief. Copies of the journal can be purchased from the Queensland Museum Shop. A Guide to Authors is displayed at the Queensland Museum web site www.qm.qld.gov.au/organisation/publications/memoirs/guidetoauthors.pdf A Queensland Government Project Typeset at the Queensland Museum THE STUDY OF TURBELLARIANS (PLATYHELMINTHES) AT THE QUEENSLAND MUSEUM B.M. ANGUS Angus, B.M. 2007 11 30: The study of turbellarians (Platyhelminthes) at the Queensland Museum. Memoirs of the Queensland Museum 53(1): 157-185. Brisbane. ISSN 0079-8835. Turbellarian research was largely ignored in Australia, apart from some early interest at the turn of the 19th century. The modern study of this mostly free-living branch of the phylum Platyhelminthes was led by Lester R.G. Cannon of the Queensland Museum. A background to the study of turbellarians is given particularly as it relates to the efforts of Cannon on symbiotic fauna, and his encouragement of visiting specialists and students. -

(Platyhelminthes, Turbellaria, Polycladida) from Karachi Coast
International Journal of Research Studies in Zoology (IJRSZ) Volume 2, Issue 2, 2016, PP 23-28 ISSN 2454-941X http://dx.doi.org/10.20431/2454-941X.0202005 www.arcjournals.org Short Notes on Marine Polycladids (Platyhelminthes, Turbellaria, Polycladida) from Karachi Coast Quddusi B. Kazmi Marine Reference Collection and Resource Centre, University of Karachi, Pakistan Abstract: Ten new records of marine polycladid worms are subject of the present notes from Pakistan. Each species is photographed and discussed briefly. 1. INTRODUCTION The Polycladida represents a highly diverse clade of free-living marine turbellarian flatworms. They are known from the littoral to the sub littoral zone. Although not related to molluscs, they are often mistaken for sea slugs because of their brilliant colour patterns. There is little known about the biodiversity of polycladid flatworms from the Indian Ocean. In Pakistan, studies on polycladids have remained neglected, first report was by Kazmi (1996), then Fatima and Barkati (1999) as Stylochoplanapallida reported Emprosthopharynxpallida (Quatrefage,1845) and latelyKazmi and Naushaba (2013) listed 4 unidentified species or only identified to genus level, of these , their unspecified genus Pseudocerosis now identified as belonging to Pseudocerossusanae Newman and Anderson ,1997 ,an undetermined pseudocertid is now named as Tytthosoceroslizardensis Newman and Cannon,1996 and another undetermined genus is given as Cestoplanarubrocinta (Grube, 1840) ,more species are added here;all are briefly described here -

Neotype Designation for the Marine Flatworm, Acanthozoon Alderi
Zoological Studies 57: 45 (2018) doi:10.6620/ZS.2018.57-45 Open Access Neotype Designation for the Marine Flatworm, Acanthozoon alderi (Polycladida: Cotylea: Pseudocerotidae), from India with Comments on the Taxonomical Status of the Genus Sudhanshu Dixit1, Verónica N. Bulnes2,*, and Chelladurai Raghunathan3 1Centre for Marine Living Resources and Ecology, Kochi, India. E-mail: [email protected] 2Laboratorio de Zoología de Invertebrados I. INBIOSUR-CONICET-UNS. San Juan 670. 8000 Bahía Blanca. Buenos Aires. Argentina 3Zoological Survey of India, M - Block, Alipore, Kolkata, India. E-mail: [email protected] (Received 16 April 2018; Accepted 16 August 2018; Published 27 September 2018; Communicated by Benny K.K. Chan) Citation: Dixit S, Bulnes VN, Raghunathan C. 2018. Neotype designation for the marine flatworm, Acanthozoon alderi (Polycladida: Cotylea: Pseudocerotidae), from India with comments on the taxonomical status of the genus. Zool Stud 57:45. doi:10.6620/ZS.2018.57- 45. Sudhanshu Dixit, Verónica N. Bulnes, and Chelladurai Raghunathan (2018) Acanthozoon alderi is an ovoid, medium-sized pseudocerotid. Body margin ruffled; pseudotentacles black and pointed, with white tips. Dorsal surface covered with papillae, except for the cerebral region. Background colour light brown, with marbled blackish pattern, middorsal black band with white blotches; black submarginal band and marginal white rim. This species was described from Borneo; however, no type specimen was designated or deposited in any museum by the author. Many nomenclature problems and misidentification have been encountered with this species (it has been identified as Acanthozoon sp. in many instances). Thus, it is necessary to designate a neotype to solve the problems of doubtful and confusing identities and maintain nomenclature stability. -

Marine-Flatworms-Of-The-Tropical-Indo-Pacific-Look
Marine Flatworms of the Tropical Indo-Pacific Photographic guide on marine polyclads with 580+ species Andrey Ryanskiy PICTORIAL INDEX TO POLYCLAD FAMILIES AND GENERA PSEUDOCEROTIDAE: ACANTHOZOON - 9 THYSANOZOON - 14 BULACEROS - 16 MAIAZOON - 17 NYMPHOZOON - 17 PHRIKOCEROS - 18 PSEUDOBICEROS - 21 PSEUDOCEROS - 38 EURYLEPTIDAE: CYCLOPORUS - 86 EURYLEPTA - 92 EURYLEPTID - 101 STYLOSTOMUM - 101 DIPOSTHIDAE: PROSTHIOSTOMIDAE: MARITIGRELLA - 102 PROSTHECERAEUS - 105 PERICELIS - 106 ENCHIRIDIUM - 107 CESTOPLANIDAE: BONINIIDAE: LURYMARE - 107 PROSTHIOSTOMUM - 112 CESTOPLANA - 112 BONINIA - 113 PLANOCERIDAE: CALLIOPLANIDAE: PARAPLANOCERA - 115 PLANOCERA - 120 CALLIOPLANA - 121 HOPLOPLANIDAE - 122 STYLOCHIDAE: GNESIOCEROTIDAE: LIMNOSTYLOCHIDAE - 122 ILYELLA - 123 STYLOCHID - 123 ECHINOPLANA - 126 NOTOPLANIDAE: UNIDENTIFIED ACOTYLEANS3 GNESIOCEROS - 126 NOTOPLANA - 127 LEPTOPLANIDAE - 128 - 129 FLATWORMS: BASIC KNOWLEDGE Why are they flat? Polyclads are considered the most primitive bilaterally symmetrical animals (left side mirrors the right). They evolved from hydra-like animals about 550 million years ago. Flatworms have no body cavity other than the gut. Respiratory and blood vessel systems are completely missing and diffusion is used for transport of oxygen inside the body. This constrains flatworms to be flat as possible for maintaining metabolism, since no cell can be too far from the outside, making a flattened body shape necessary. How do they eat? Flatworms/Polyclads have a mouth with pharynx inside: a muscular tube through which the flatworm can suck food. Pharynx may be tubular or ruffled with numerous folds (more details on External Morphology Basics pages) Flatworms are carnivorous, feeding on small invertebrates, suctioning entirely their prey or digesting a part of it. Many species of the Pseudocerotidae family prefer ascidians, sponges, and bryozoans. For feeding, the pharynx protrudes and can be expanded into the individual zooids of colonial ascidians. -

From Cape Verde and Related Regions of Macaronesia
European Journal of Taxonomy 736: 1–43 ISSN 2118-9773 https://doi.org/10.5852/ejt.2021.736.1249 www.europeanjournaloftaxonomy.eu 2021 · Cuadrado D. et al. This work is licensed under a Creative Commons Attribution License (CC BY 4.0). Research article urn:lsid:zoobank.org:pub:FC9085BE-73C4-4F33-BD9B-6A9F573AB01D Polycladida (Platyhelminthes, Rhabditophora) from Cape Verde and related regions of Macaronesia Daniel CUADRADO 1, Jorge RODRÍGUEZ 2, Leopoldo MORO 3, Cristina GRANDE 4 & Carolina NOREÑA 5,* 1,5 Departmento de Biodiversidad y Biología Evolutiva, Museo Nacional de Ciencias Naturales (CSIC), c/ José Gutiérrez Abascal 2, 28006 Madrid, Spain. 2 Marine Invertebrates Department, Australian Museum Research Institute, Australian Museum, 1 William Street, Sydney, NSW 2010, Australia. 3 Servicio de Biodiversidad, Gobierno de Canarias, Edif. Usos Múltiples I, Av. Anaga n° 35, Pl. 11, 38071 S/C de Tenerife, Canary Islands, Spain. 4 Departamento de Biología, Facultad de Ciencias, Universidad Autónoma de Madrid, Cantoblanco, 28049 Madrid, Spain. * Corresponding author: [email protected] 1 Email: [email protected] 2 Email: [email protected] 3 Email: [email protected] 4 Email: [email protected] 1 urn:lsid:zoobank.org:author:F0C14D94-9996-4A20-9D56-B02DDA1A78CA 2 urn:lsid:zoobank.org:author:B833502E-CBA4-40CA-AE5A-BAD02F539062 3 urn:lsid:zoobank.org:author:B66DDDE6-98E6-42FD-8E58-A1DF6A386BE5 4 urn:lsid:zoobank.org:author:C8634A50-D3EC-467A-A868-225C231B40F2 5 urn:lsid:zoobank.org:author:DD03B71F-B45E-402B-BA32-BB30343E0D95 Abstract. The systematics and distribution of the order Polycladida within the Macaronesian archipelagos are analysed. New species (Marcusia alba sp. -
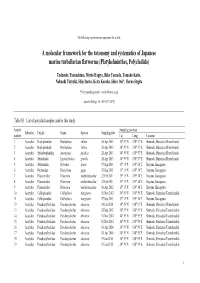
Platyhelminthes, Polycladida)
The following supplement accompanies the article A molecular framework for the taxonomy and systematics of Japanese marine turbellarian flatworms (Platyhelminthes, Polycladida) Tadasuke Tsunashima, Morio Hagiya, Riko Yamada, Tomoko Koito, Nobuaki Tsuyuki, Shin Izawa, Keita Kosoba, Shiro Itoi*, Haruo Sugita *Corresponding author: [email protected] Aquatic Biology 26: 159–167 (2017) Table S1. List of polyclad samples used in this study Sample Sampling location Suborder Family Genus Species Sampling date number Lat. Long. Location 1 Acotylea Hoploplanidae Hoploplana villosa 26 Apr 2013 34° 39' N 138° 57' E Shimoda, Shizuoka (Ebisu Island) 2 Acotylea Hoploplanidae Hoploplana villosa 26 Apr 2013 34° 39' N 138° 57' E Shimoda, Shizuoka (Ebisu Island) 3 Acotylea Stylochoplanidae Amemiyaia pacifica 26 Apr 2013 34° 39' N 138° 57' E Shimoda, Shizuoka (Ebisu Island) 4 Acotylea Stylochidae Leptostylochus gracilis 26 Apr 2013 34° 39' N 138° 57' E Shimoda, Shizuoka (Ebisu Island) 5 Acotylea Stylochidae Stylochus ijimai 07 Sep 2013 35° 15' N 139° 34' E Hayama, Kanagawa 6 Acotylea Ilyplanidae Discoplana gigas 07 Sep 2013 35° 15' N 139° 34' E Hayama, Kanagawa 7 Acotylea Planoceridae Planocera multitentaculata 22 Feb 2011 35° 15' N 139° 34' E Hayama, Kanagawa 8 Acotylea Planoceridae Planocera multitentaculata 22 Feb 2011 35° 15' N 139° 34' E Hayama, Kanagawa 9 Acotylea Planoceridae Planocera multitentaculata 06 Apr 2012 35° 15' N 139° 34' E Hayama, Kanagawa 10 Acotylea Callioplanidae Callioplana marginata 01 Nov 2013 34° 39' N 138° 59' E Shimoda, Shizuoka -

Pseudoceros Astrorum, a New Species of Polycladida (Cotylea, Pseudocerotidae) from Northeastern Brazil
Zootaxa 3881 (1): 094–100 ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2014 Magnolia Press ISSN 1175-5334 (online edition) http://dx.doi.org/10.11646/zootaxa.3881.1.7 http://zoobank.org/urn:lsid:zoobank.org:pub:9C0FB9F8-C565-4627-87A2-D44FDFAB6781 Pseudoceros astrorum, a new species of Polycladida (Cotylea, Pseudocerotidae) from Northeastern Brazil VERONICA N. BULNES1 & YAN TORRES2 1Zoología de Invertebrados I, Universidad Nacional del Sur. San Juan 670. 8000 Bahía Blanca, Argentina 2Departamento de Biologia, Centro de Ciências, Universidade Federal do Ceará, Ceará, Brasil Abstract Pseudoceros astrorum n. sp. is characterized by a smooth dorsal surface with a brown ground colour, and with net-like pattern of small black granules, white spots of different sizes uniformly distributed, a thin black sub-marginal band, and a white marginal rim. The pseudotentacles are dark brown with white tips and the anterior margin and cerebral region is devoid of pigmentation. The male system is characterised by conspicuous spermiducal bulbs, a conical curved penis stylet, and the sucker lies more or less posterior. With this contribution, the number of known species from Brazil is now 72, and has created new interest in the lesser-known polyclad fauna from the northeast coast of Brazil. Key words: biodiversity, morphology, Marine flatworms, Western Atlantic, Cotylea Introduction The Pseudocerotidae is one of the largest families within the Cotylea, with a large number of described species. The phylogeny as well as the monophyly of some genera are still unclear (Rawlinson & Litvaitis 2008). Pseudoceros is one of the most common tropical polyclad genera, and species of Pseudoceros are determined on the basis of their colour patterns (Newman & Cannon 1995), external morphological features, as well as the reproductive biology, and microanatomy (Hyman 1951; Prudhoe 1985; Faubel 1984; Bolaños et al. -

State of Kent's Wildlife
TheKent’s State of Wildlife in 2011 Kent Biodiversity Partnership Action for Kent’s wildlife Contents Introduction 1 Kent’s Butterflies Mike Easterbrook Butterfly Conservation -Kent 2 Kent’s Moths Ian Ferguson & David Gardner Butterfly Conservation -Kent 5 Kent’s Amphibians and Reptiles Dr Lee Brady Kent Reptile and Amphibian Group 10 Kent’s Birds Andrew Henderson Kent Ornithological Society 18 Kent’s Bats Shirley Thompson Kent Bat Group 26 Kent’s Wild Plants Richard Moyse Kent Wildlife Trust 33 Kent’s Marine Wildlife Bryony Chapman, Kent Wildlife Trust 39 The State of Kent’s Wildlife in 2011 Kent is one the UK’s most wildlife-rich bird species, and two species of bat all counties, a result of its varied geology, became extinct in the county. This long coastline, landscape history, excludes consideration of groups not southerly location and proximity to covered in the following chapters; for mainland Europe. Its important wildlife example, the Red Squirrel and 3 species habitats include estuaries, chalk cliffs, of bumblebee were also lost during the woodlands, and chalk downland, and 20th century. In addition to this, many of encompass some of the South East’s the species that remain have seen big most iconic landscapes, such as the population declines, including many shingle headland of Dungeness and species of butterflies and moths, birds the White Cliffs of Dover. and wildflowers of farmland, wetland plants, Adders and Common Toads. This publication has been prepared by Kent natural historians to give an As seen in the following chapters, the outline of the changing fortunes of causes of these losses and declines are Kent’s wild plants and animals over the various. -

Newman Et Al 2003
Micronesica 35-36:189-199. 2003 Checklist of polyclad flatworms (Platyhelminthes) from Micronesian coral reefs L. J. NEWMAN School of Environmental Science & Management Southern Cross University PO Box 157 Lismore, NSW Australia 2480 email:[email protected] G. PAULAY1, R. RITSON-WILLIAMS2 Marine Laboratory University of Guam Mangilao, Guam 96923 U.S.A AbstractWe record 68 species of polyclad flatworms from new material (all photo-documented) and 28 species from literature records, for a total diversity of 88 species for Micronesia. Up to 60% of the encountered species may be undescribed. Guam has the largest recorded fauna with 59 species, followed by 28 species known from Palau. Pseudocerotidae comprise 58% of documented species, and more than 3 times as many cotyleans than acotyleans are documented. This study shows that the polyclad fauna of Micronesia is diverse yet poorly known, and highlights the need for further work. Introduction Polyclad flatworms are conspicuous inhabitants of coral reefs especially throughout the Indo-West Pacific, yet their diversity within Micronesia remains poorly documented. Only five papers have dealt with the polyclad fauna of this large and diverse area (Kato 1943, Hyman 1955, 1959; Newman & Cannon 1997, Newman & Schupp 2002). This checklist represents the first comprehensive account of polyclad flatworms from Micronesian waters. Although many tropical polyclads are brightly colored and attract attention, they remain understudied partly for methodological reasons. Accurate taxonomic determinations involve examination of both the morphology of living animals and the anatomy of the reproductive structures (Newman & Cannon 1994a). Diagnostic color characters tend to disappear rapidly after fixation and need to be documented photographically from living animals. -

STUDIES on the FAUNA of CURAÇAO and OTHER CARIBBEAN ISLANDS: No
STUDIES ON THE FAUNA OF CURAÇAO AND OTHER CARIBBEAN ISLANDS: No. 101. Polycladida from Curaçao and faunistically related regions by Eveline du Bois-Reymond Marcus & Ernst Marcus (Departamento de Zoologia, Universidade de Sao Paulo) Professor Dr. DIVA DINIZ CORRÊA, Head of the Department of Zoology of the University of São Paulo, was able to work at the “Caraïbisch Marien-Biologisch Instituut” (Caribbean Marine Bio- logical Institute: Carmabi) at Curaçao from December 1965 to March 1966, thanks to a grant received from the Government of the Netherlands. There she collected 26 species of Polyclads, and took notes of their shapes and colours. Furthermore Dr. PIETER WAGENAAR HUMMELINCK, of Utrecht, Caribbean sent us a large collection of polyclads from the area, from He had also collected in gathered on his trips 1930 to 1964. 1963 in the Miami area. We received some samples from the latter area from Prof. Dr. CORRÊA and Prof. Dr. FREDERICK M. BAYER, of Miami. Drs. LILIANA FORNERIS, WALTER NARCHI, and SÉRGIO DE ALMEIDA all of São Brazilian RODRIGUES, Paulo, gave us interesting material. The indications (B), (C), (F), (H), or (N) after the date denote the collectors. Dr. HUMMELINCK'S station numbers (H 1008A, 1064b, under of the be etc., cf. Fig. 100), which a description habitat can found, refer to his list of 1930-1949 localities in the 4th volume of this series, or to a forthcoming paper, in which the 1955-1964 lo- calities will be described. The specimens collected by Dr. HUMMELINCK and Dr. BAYER will be returned to Utrecht and Miami, respectively, the other ones are kept in the Department of Zoology, Faculty of Philosophy, University of Sao Paulo.