Characterization of a Full-Length Infectious Clone of Bovine Foamy Virus 3026
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Foamy Virus Assembly with Emphasis on Pol Encapsidation
Viruses 2013, 5, 886-900; doi:10.3390/v5030886 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Foamy Virus Assembly with Emphasis on Pol Encapsidation Eun-Gyung Lee 1, Carolyn R. Stenbak 2 and Maxine L. Linial 1,* 1 Fred Hutchinson Cancer Research Center, Basic Sciences Division; 1100 Fairview Avenue North, Seattle, WA 98109, USA; E-Mails: [email protected] (EGL); [email protected] (MLL) 2 Seattle University, Biology Department; 901 12th Avenue, Seattle, WA 98122, USA; E-Mail: [email protected] * Authors to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-206- 667-4442; Fax: +1-206-667-5939 Received: 31 January 2013; in revised form: 11 March 2013 / Accepted: 14 March 2013 / Published: 20 March 2013 Abstract: Foamy viruses (FVs) differ from all other genera of retroviruses (orthoretroviruses) in many aspects of viral replication. In this review, we discuss FV assembly, with special emphasis on Pol incorporation. FV assembly takes place intracellularly, near the pericentriolar region, at a site similar to that used by betaretroviruses. The regions of Gag, Pol and genomic RNA required for viral assembly are described. In contrast to orthoretroviral Pol, which is synthesized as a Gag-Pol fusion protein and packaged through Gag-Gag interactions, FV Pol is synthesized from a spliced mRNA lacking all Gag sequences. Thus, encapsidation of FV Pol requires a different mechanism. We detail how WT Pol lacking Gag sequences is incorporated into virus particles. In addition, a mutant in which Pol is expressed as an orthoretroviral-like Gag-Pol fusion protein is discussed. -

D3.7 First Periodic Report on Ongoing Jrps WP3 Joint Research Projects
D3.7 First periodic report on ongoing JRPs WP3 Joint Research Projects Responsible Partner: Sciensano Contributing partners: / 1 GENERAL INFORMATION European Joint Programme Promoting One Health in Europe through joint actions on foodborne full title zoonoses, antimicrobial resistance and emerging microbiological hazards European Joint Programme One Health EJP acronym Funding This project has received funding from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No 773830. Grant Agreement Grant agreement n° 773830 Starting Date 01/01/2018 Duration 60 Months DOCUMENT MANAGEMENT Deliverable D3.7 First periodic report on ongoing JRPs WP and Task WP3; Task 3.2 Leader Sciensano Other contributors ANSES, SVA, WBVR, INSA Due month of the deliverable M13 Actual submission month M14 Type R R: Document, report DEC: Websites, patent fillings, videos, etc. OTHER Dissemination level PU PU: Public CO: confidential, only for members of the consortium (including the Commission Services) First periodic report on ongoing JRP Introduction Joint Research Projects serve as an instrument that help OneHealth EJP partners to work together in developing new detection methods, improved diagnostic tests, fast and accurate typing methods, in gaining new insight in the spread of pathogens and their resistance traits etc. At the same time, through setting up these scientific collaborations, researchers spread over Europe identify new possible partners and strengthen links between known colleagues. As such, the JRP help in creating and consolidating a firm network of organizations that have reference tasks in their scope and that deal with foodborne zoonoses, antimicrobial resistance and emerging threats. Summary of the performance of the Joint Research Projects Project deliverables and milestones The 11 joint research projects planned to submit a total of 77 deliverables. -

Eukaryote Cell Biology - Michelle Gehringer
FUNDAMENTALS OF BIOCHEMISTRY, CELL BIOLOGY AND BIOPHYSICS – Vol. II - Eukaryote Cell Biology - Michelle Gehringer EUKARYOTE CELL BIOLOGY Michelle Gehringer Department of Biochemistry and Microbiology, University of Port Elizabeth, South Africa Keywords: cell theory, cell diversity, eukaryote cell structure, nucleus, chromatin, DNA, organelles, mitochondria, chloroplasts, transcription, RNA, translation, ribosomes, cell cycle, interphase, mitosis, meiosis, signal transduction, growth regulation, cancer, oncogenesis. Contents 1. Introduction 1.1. The first cell 2. Origin of Eukaryotes 3. Cellular differentiation in multicellular organisms 3.1. Plants 3.2. Animals 4. Eukaryotic cell structure 5. Organization of eukaryotic cells 5.1. Plasma membrane 5.2. Extracellular matrices 5.3. Protein synthesis and transport 5.4. Cytoskeleton and movement 5.5. Nucleus 5.5.1 Genomes 5.5.2 Gene expression 5.5.3 Maintaining the genome 5.6. Organelles 6. The cell cycle 6.1. Mitosis 6.2. Meiosis 7. Regulation of cell growth 7.1. Signal transduction 7.2. Programmed cell death 7.3. CancerUNESCO – EOLSS 8. Experimental Models 8.1. Yeast SAMPLE CHAPTERS 8.2. Arabidopsis 8.3. Drosophila 8.4. The mouse 8.5. Cell culture 8.6. Separation of cellular contents 8.7. Tracing biochemical pathways 9. Future Investigations Glossary Bibliography ©Encyclopedia of Life Support Systems (EOLSS) FUNDAMENTALS OF BIOCHEMISTRY, CELL BIOLOGY AND BIOPHYSICS – Vol. II - Eukaryote Cell Biology - Michelle Gehringer Biographical Sketch Summary Cells form the basic unit of life on our planet. They are well organized systems which perform all the essential tasks of eating, respiring, replicating and excreting waste products. The first cells, which are thought to have evolved about 3.8 billion years ago, much resembled present day prokaryotes. -

Characterization of Biofilm Extracts from Two Marine Bacteria
applied sciences Article Characterization of Biofilm Extracts from Two Marine Bacteria Delphine Passerini 1, Florian Fécamp 1, Laetitia Marchand 1, Laetitia Kolypczuk 1 , Sandrine Bonnetot 1, Corinne Sinquin 1 ,Véronique Verrez-Bagnis 1 , Dominique Hervio-Heath 2 , Sylvia Colliec-Jouault 1 and Christine Delbarre-Ladrat 1,* 1 Ifremer, Atlantique Center, Microbial Ecosystems and Marine Molecules for the Biotechnologies, 44311 Rue de l’Ile d’Yeu, Nantes CEDEX 3, France; [email protected] (D.P.); fl[email protected] (F.F.); [email protected] (L.M.); [email protected] (L.K.); [email protected] (S.B.); [email protected] (C.S.); [email protected] (V.V.-B.); [email protected] (S.C.-J.) 2 Ifremer, Bretagne Center, Health, Environment and Microbiology, 1625 Route de Sainte-Anne, 29280 Plouzané, France; [email protected] * Correspondence: [email protected] Received: 17 September 2019; Accepted: 13 November 2019; Published: 19 November 2019 Featured Application: By analyzing extracts of biofilm formed by two marine bacteria and comparing them with planktonic extracts, we have shown that biofilm may induce the biosynthesis of potentially bioactive compounds and may open up new possibilities for compound discovery. Abstract: In the marine environment, biofilm formation is an important lifestyle for microorganisms. A biofilm is comprised of cells embedded in an extracellular matrix that holds them close together and keeps the biofilm attached to the colonized surface. This predominant lifestyle and its main regulation pathway, namely quorum-sensing (QS), have been shown to induce specific bioactive metabolites. In this study, we investigated the biofilm formation by two marine bacteria belonging to the Vibrio species to discover potentially innovative bioactive compounds. -

The Origin of the Eukaryotic Cell Based on Conservation of Existing
The Origin of the Eukaryotic Albert D. G. de Roos The Beagle Armada Cell Based on Conservation Bioinformatics Division of Existing Interfaces Einsteinstraat 67 3316GG Dordrecht, The Netherlands [email protected] Abstract Current theories about the origin of the eukaryotic Keywords cell all assume that during evolution a prokaryotic cell acquired a Evolution, nucleus, eukaryotes, self-assembly, cellular membranes nucleus. Here, it is shown that a scenario in which the nucleus acquired a plasma membrane is inherently less complex because existing interfaces remain intact during evolution. Using this scenario, the evolution to the first eukaryotic cell can be modeled in three steps, based on the self-assembly of cellular membranes by lipid-protein interactions. First, the inclusion of chromosomes in a nuclear membrane is mediated by interactions between laminar proteins and lipid vesicles. Second, the formation of a primitive endoplasmic reticulum, or exomembrane, is induced by the expression of intrinsic membrane proteins. Third, a plasma membrane is formed by fusion of exomembrane vesicles on the cytoskeletal protein scaffold. All three self-assembly processes occur both in vivo and in vitro. This new model provides a gradual Darwinistic evolutionary model of the origins of the eukaryotic cell and suggests an inherent ability of an ancestral, primitive genome to induce its own inclusion in a membrane. 1 Introduction The origin of eukaryotes is one of the major challenges in evolutionary cell biology. No inter- mediates between prokaryotes and eukaryotes have been found, and the steps leading to eukaryotic endomembranes and endoskeleton are poorly understood. There are basically two competing classes of hypotheses: the endosymbiotic and the autogenic. -

Identification of Virus-Encoded Micrornas in Divergent Papillomaviruses
RESEARCH ARTICLE Identification of virus-encoded microRNAs in divergent Papillomaviruses Rachel Chirayil1³, Rodney P. Kincaid1³, Christine Dahlke2³, Chad V. Kuny1³, Nicole DaÈlken2, Michael Spohn2, Becki Lawson3, Adam Grundhoff2*, Christopher S. Sullivan1* 1 Center for Systems and Synthetic Biology, Center for Infectious Disease and Dept. Molecular Biosciences, The University of Texas at Austin, Austin, TX, United States of America, 2 Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg, Germany, 3 Institute of Zoology, Zoological Society of London, London, United Kingdom a1111111111 a1111111111 ³ These authors share first authorship on this work. a1111111111 * [email protected] (AG); [email protected] (CSS) a1111111111 a1111111111 Abstract MicroRNAs (miRNAs) are small RNAs that regulate diverse biological processes including multiple aspects of the host-pathogen interface. Consequently, miRNAs are commonly OPEN ACCESS encoded by viruses that undergo long-term persistent infection. Papillomaviruses (PVs) are Citation: Chirayil R, Kincaid RP, Dahlke C, Kuny capable of undergoing persistent infection, but as yet, no widely-accepted PV-encoded miR- CV, DaÈlken N, Spohn M, et al. (2018) Identification of virus-encoded microRNAs in divergent NAs have been described. The incomplete understanding of PV-encoded miRNAs is due in Papillomaviruses. PLoS Pathog 14(7): e1007156. part to lack of tractable laboratory models for most PV types. To overcome this, we have https://doi.org/10.1371/journal.ppat.1007156 developed miRNA Discovery by forced Genome Expression (miDGE), a new wet bench Editor: Paul Francis Lambert, University of approach to miRNA identification that screens numerous pathogen genomes in parallel. Wisconsin Madison School of Medicine and Public Using miDGE, we screened over 73 different PV genomes for the ability to code for miRNAs. -

Molecular Epidemiology and Whole-Genome Analysis of Bovine Foamy Virus in Japan
viruses Article Molecular Epidemiology and Whole-Genome Analysis of Bovine Foamy Virus in Japan Hirohisa Mekata 1,* , Tomohiro Okagawa 2, Satoru Konnai 2,3 and Takayuki Miyazawa 4 1 Center for Animal Disease Control, University of Miyazaki, Miyazaki 889-2192, Japan 2 Department of Advanced Pharmaceutics, Faculty of Veterinary Medicine, Hokkaido University, Sapporo 060-0818, Japan; [email protected] (T.O.); [email protected] (S.K.) 3 Department of Disease Control, Faculty of Veterinary Medicine, Hokkaido University, Sapporo 060-0818, Japan 4 Laboratory of Virus-Host Coevolution, Institute for Frontier Life and Medical Sciences, Kyoto University, Kyoto 606-8507, Japan; [email protected] * Correspondence: [email protected]; Tel./Fax: +81-985-58-7881 Abstract: Bovine foamy virus (BFV) is a member of the foamy virus family in cattle. Information on the epidemiology, transmission routes, and whole-genome sequences of BFV is still limited. To understand the characteristics of BFV, this study included a molecular survey in Japan and the determination of the whole-genome sequences of 30 BFV isolates. A total of 30 (3.4%, 30/884) cattle were infected with BFV according to PCR analysis. Cattle less than 48 months old were scarcely infected with this virus, and older animals had a significantly higher rate of infection. To reveal the possibility of vertical transmission, we additionally surveyed 77 pairs of dams and 3-month-old calves in a farm already confirmed to have BFV. We confirmed that one of the calves born from a dam with BFV was infected. -

Sensitivity of Golgi Matrix Proteins to an ER Exit Block
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central JCBArticle Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block Suzanne Miles,1 Heather McManus,1 Kimberly E. Forsten,2 and Brian Storrie1 1Department of Biochemistry and 2Department of Chemical Engineering, Virginia Tech, Blacksburg, VA 24061 e tested whether the entire Golgi apparatus is concentration. Redistribution of GalNAcT2 was more a dynamic structure in interphase mammalian sensitive to low Sar1pdn concentrations than giantin or Wcells by assessing the response of 12 different GM130. Redistribution was most rapid for p27, COPI, and Golgi region proteins to an endoplasmic reticulum (ER) exit p115. Giantin, GM130, and GalNAcT2 relocated with block. The proteins chosen spanned the Golgi apparatus approximately equal kinetics. Distinct ER accumulation and included both Golgi glycosyltransferases and putative could be demonstrated for all integral membrane proteins. matrix proteins. Protein exit from ER was blocked either by ER-accumulated Golgi region proteins were functional. microinjection of a GTP-restricted Sar1p mutant protein in Photobleaching experiments indicated that Golgi-to-ER the presence of a protein synthesis inhibitor, or by plasmid- protein cycling occurred in the absence of any ER exit encoded expression of the same dominant negative Sar1p. block. We conclude that the entire Golgi apparatus is a All Golgi region proteins examined lost juxtanuclear Golgi dynamic structure and suggest that most, if not all, Golgi apparatus–like distribution as scored by conventional and region–integral membrane proteins cycle through ER in confocal fluorescence microscopy in response to an ER interphase cells. -

10 International Foamy Virus Conference 2014
10th INTERNATIONAL FOAMY VIRUS CONFERENCE 2014 June 24‐25, 2014 National Veterinary Research Institute, Pulawy, Poland Organized by: National Veterinary Research Institute Committee of Veterinary Sciences Polish Academy of Sciences Topics include: Foamy Virus Infection and Zoonotic Aspects, Foamy Virus Restriction and Immunity, Foamy Virus Biology and Gene Therapy Vectors http://www.piwet.pulawy.pl/ifvc/ Organizers Jacek Kuźmak and Magdalena Materniak Scientific Advisors Martin Löchelt and Axel Rethwilm Sponsors Support for the meeting was provided by the Committee of Veterinary Sciences Polish Academy of Sciences, ROCHE, Thermo Fisher Scientific and EURx Abstract book was issued thanks to the support provided by Committee of Veterinary Sciences Polish Academy of Sciences 2 Program Overview Monday, June 23rd, 2014 19 00 – 20 00 Registration 20 30 – open end Welcome Reception Tuesday, June 24th, 2014 9 00 – 9 30 Registration 9 30 – 9 45 Welcome 9 45 – 10 15 Arifa Khan (FDA, Bethesda, US) - Simian foamy virus infection in natural host species 10 15 – 10 45 Antoine Gessain (Institute Pasteur, Paris, France) – Zoonotic human infection by simian foamy viruses 10 45 – 11 20 Session 1 on Foamy Virus Infection and Zoonotic Aspects Chairs: Arifa S. Khan and Antoine Gessain 11 20 – 11 45 Coffee break 11 45 – 12 30 Session 1 on Foamy Virus Infection and Zoonotic Aspects Chairs: Arifa S. Khan and Antoine Gessain 12 30 – 13 00 Aris Katzourakis (Oxford University, UK - Mammalian genomes, their endogenous viral elements, and the evolutionary history -

Center of Biomedical Research Excellence in Matrix Biology: Building Research Infrastructure, Supporting Young Researchers, and Fostering Collaboration
International Journal of Molecular Sciences Editorial Center of Biomedical Research Excellence in Matrix Biology: Building Research Infrastructure, Supporting Young Researchers, and Fostering Collaboration Julia Thom Oxford 1,2,3,* , Ken A. Cornell 1,3,4, Jared J. Romero 1,5 , Diane B. Smith 1,3 , Tracy L. Yarnell 1,3, Rhiannon M. Wood 1,3 , Cheryl L. Jorcyk 1,2, Trevor J. Lujan 1,6, Allan R. Albig 1,2 , Kristen A. Mitchell 1,2, Owen M. McDougal 1,4 , Daniel Fologea 1,7, David Estrada 1,8, Juliette K. Tinker 1,2, Rajesh Nagarajan 1,4, Don L. Warner 1,4, Troy T. Rohn 1,2, Jim Browning 1,9 , Richard S. Beard Jr. 1,3, Lisa R. Warner 1,4 , Brad E. Morrison 1,2, Clare K. Fitzpatrick 1,6, Gunes Uzer 1,6, Laura Bond 1,3, Stephanie M. Frahs 1,3, Cynthia Keller-Peck 1,3, Xinzhu Pu 1,3, Luke G. Woodbury 1,3 and Matthew W. Turner 1,3 1 Center of Biomedical Research Excellence in Matrix Biology, Boise State University, Boise, ID 83725, USA 2 Department of Biological Sciences, Boise State University, Boise, ID 83725, USA 3 Biomolecular Research Center, Boise State University, Boise, ID 83725, USA 4 Department of Chemistry and Biochemistry, Boise State University, Boise, ID 83725, USA 5 Division of Research, Boise State University, Boise, ID 83725, USA 6 Department of Mechanical and Biomedical Engineering, Boise State University, Boise, ID 83725, USA 7 Department of Physics, Boise State University, Boise, ID 83725, USA 8 Micron School of Materials Science and Engineering, Boise State University, Boise, ID 83725, USA 9 Department of Electrical and Computer Engineering, Boise State University, Boise, ID 83725, USA * Correspondence: [email protected]; Tel.: +01-208-426-2395 Received: 9 March 2020; Accepted: 17 March 2020; Published: 20 March 2020 Abstract: The Center of Biomedical Research Excellence in Matrix Biology strives to improve our understanding of extracellular matrix at molecular, cellular, tissue, and organismal levels to generate new knowledge about pathophysiology, normal development, and regenerative medicine. -
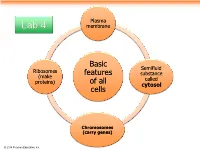
Basic Features of All Cells
Plasma Lab 4 membrane Basic Semifluid Ribosomes features substance (make called proteins) of all cytosol cells Chromosomes (carry genes) © 2014 Pearson Education, Inc. Prokaryotic cells are characterized by having . No nucleus . DNA in an unbound region called the nucleoid . No membrane-bound organelles . Cytoplasm bound by the plasma membrane © 2014 Pearson Education, Inc. Eukaryotic cells are characterized by having • DNA in a nucleus that is bounded by a membranous nuclear envelope • Membrane-bound organelles • Cytoplasm in the region between the plasma membrane and nucleus Eukaryotic cells are generally much larger than prokaryotic cells © 2014 Pearson Education, Inc. Figure 6.8a ENDOPLASMIC RETICULUM (ER) Nuclear envelope Rough ER Smooth ER Nucleolus NUCLEUS Flagellum Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Lysosome Mitochondrion © 2014 Pearson Education, Inc. Figure 6.8b Nuclear envelope NUCLEUS Nucleolus Rough ER Chromatin Smooth ER Ribosomes Golgi Central vacuole apparatus Microfilaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Plasma Chloroplast membrane Cell wall Plasmodesmata Wall of adjacent cell © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. Table 6.1 © 2014 Pearson Education, Inc. Cell Walls of Plants The cell wall is an extracellular structure that distinguishes plant cells from animal cells Plant cell walls Prokaryotes, are made -

An Endogenous Foamy-Like Viral Element in the Coelacanth Genome
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central An Endogenous Foamy-like Viral Element in the Coelacanth Genome Guan-Zhu Han*, Michael Worobey* Department of Ecology and Evolutionary Biology, University of Arizona, Tucson, Arizona, United States of America Abstract Little is known about the origin and long-term evolutionary mode of retroviruses. Retroviruses can integrate into their hosts’ genomes, providing a molecular fossil record for studying their deep history. Here we report the discovery of an endogenous foamy virus-like element, which we designate ‘coelacanth endogenous foamy-like virus’ (CoeEFV), within the genome of the coelacanth (Latimeria chalumnae). Phylogenetic analyses place CoeEFV basal to all known foamy viruses, strongly suggesting an ancient ocean origin of this major retroviral lineage, which had previously been known to infect only land mammals. The discovery of CoeEFV reveals the presence of foamy-like viruses in species outside the Mammalia. We show that foamy-like viruses have likely codiverged with their vertebrate hosts for more than 407 million years and underwent an evolutionary transition from water to land with their vertebrate hosts. These findings suggest an ancient marine origin of retroviruses and have important implications in understanding foamy virus biology. Citation: Han G-Z, Worobey M (2012) An Endogenous Foamy-like Viral Element in the Coelacanth Genome. PLoS Pathog 8(6): e1002790. doi:10.1371/ journal.ppat.1002790 Editor: Michael Emerman, Fred Hutchinson Cancer Research Center, United States of America Received February 29, 2012; Accepted May 22, 2012; Published June 28, 2012 Copyright: ß 2012 Han, Worobey.