Deciphering Biodiversity and Interactions Between
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Reconstruction Du Pont Sur La Loue À Chamblay Demande De Dérogation Pour La Destruction, L'altération, Ou La Dégradation D
Reconstruction du pont sur la Loue à Chamblay Demande de dérogation pour la destruction, l’altération, ou la dégradation de sites de reproduction et aires de repos d’espèces animales protégées PMM/ Cabinet REILE Pascal - année 2013 __________________________________ -1/46- PMM/ Cabinet REILE Pascal - année 2013 __________________________________ -2/46- Reconstruction du pont sur la Loue à Chamblay Note complémentaire Reconstruction du pont sur la Loue à Chamblay Demande de dérogation pour la destruction, l’altération, ou la dégradation de sites de reproduction et aires de repos d’espèces animales protégées Octobre 2013 Le Conseil Général du Jura projette la reconstruction du pont sur la Loue à Chamblay. Ce projet et les travaux annexes vont toucher des habitats, plus précisément des zones de reproduction (supports de nids et modification d’une mare) de Bergeronnette grise, de Rougequeue noir, et de Grenouille verte. S’agissant d’espèces protégées, la modification de leur habitat est soumise à demande de dérogation en application de l’article L.411-2 du Code de l’Environnement portant sur des espèces de faune et de flore sauvage protégées. La présente note est la demande de dérogation du maitre d’ouvrage du projet, le conseil Général du Jura : Le CONSEIL GENERAL DU JURA Direction des Equipements Départementaux et de leur Maintenance 17, rue Rouget de l’Isle 39 039 LONS LE SAUNIER Comme demandé dans l’arrêté du 19 février 2007 fixant les conditions d’instruction de ce type de demande, ce document présente l’inventaire phyto-écologique réalisé sur le site, les espèces concernées, une justification des travaux, et les mesures compensatoires proposées par le maitre d’ouvrage. -

N° 7-2 Juillet 2009
N° 7-2 PREFECTURE DU JURA JUILLET 2009 I.S.S.N. 0753 - 4787 Papier écologique 8 RUE DE LA PREFECTURE - 39030 LONS LE SAUNIER CEDEX - : 03 84 86 84 00 - TELECOPIE : 03 84 43 42 86 - INTERNET : www.jura.pref.gouv.fr 566 DIRECTION REGIONALE DE L’INDUSTRIE, DE LA RECHERCHE ET DE L’ENVIRONNEMENT DE FRANCHE-COMTE .........................................................................................................................................................................................................................567 Arrêté n° 09-123 du 13 juillet 2009 portant subdélégation de signature ................................................................................................567 DIRECTION DEPARTEMENTALE DE L’EQUIPEMENT ET DE L’AGRICULTURE .....................................................................569 Commission départementale de la chasse et de la faune sauvage - Formation spécialisée dégâts de gibier - Compte rendu de la réunion du 10 juillet 2009 .....................................................................................................................................................................................569 Arrêté préfectoral n° 2009/472 du 29 juin 2009 portant création d’une réserve de chasse et de faune sauvage sur le territoire de l’ACCA de VILLERS ROBERT ................................................................................................................................................................570 Arrêté préfectoral n° 2009/473 du 29 juin 2009 portant création d’une -
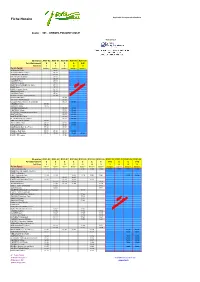
Fiche Horaire Applicable À Compter Du 01/09/2016
Fiche Horaire Applicable à compter du 01/09/2016 Ligne : 301 - ARBOIS-POLIGNY-DOLE Transporteur : Itinéraires A301-02 A301-01 A301-03 A301-07 A301-09 Fonctionnement S S S A V+E Services 5 1 3 15 19 Point d'arrêt lmmjv--- lmmjv--- lmmjv--- lmmjv--- lmmjv--- ARBOIS La Foule 06:45 ARBOIS Arbois Tourisme 06:48 VADANS Carf. RD 469 06:52 MATHENAY Molamboz 06:54 FERTE (LA) Chalet 06:57 VAUDREY Pont 07:03 VAUDREY Eglise 07:05 MONT S/S VAUDREY Car. Gare 07:11 BANS Mairie 07:13 BANS Rue des Tilleuls 07:14 SOUVANS RN 63 07:15 SOUVANS Poste 07:16 NEVY LES DOLE Place Bascule 07:20 MOUCHARD Mairie 06:45 VILLERS FARLAY Mairie 06:50 ECLEUX Place Monument aux Morts 06:53 13:02 CHAMBLAY Mairie 06:50 OUNANS Eglise 06:55 CHISSEY/LOUE Ecole 06:58 13:04 CHATELAY Village 07:00 13:06 GERMIGNEY Monuments aux Morts 07:04 13:10 SANTANS Place 07:07 13:13 MONTBARREY Place 07:09 13:15 BELMONT Rue Val d'Amour 07:13 13:19 VIEILLE LOYE (LA) Mairie 07:04 AUGERANS Place 07:12 07:15 13:21 LOYE (LA) Place 07:20 07:24 13:30 DOLE/GOUX Salle des Fêtes 07:24 07:29 DOLE/GOUX Mairie 07:27 DOLE Le Port-Stade 07:31 07:35 07:33 13:39 DOLE Gare Routière 07:36 07:40 07:36 13:45 07:46 DOLE CES Ledoux 07:50 Itinéraires R301-03 R301-02 R301-07 R301-04 R301-05 R301-03 R301-06 R301-10 R301-11 R301-08 R301-08 Fonctionnement S S S S S S S V+E S S V+E Services 8 4 20 12 14 18 16 22 24 10 26 Point d'arrêt --m----- --m----- --m----- lm-jv--- lm-jv--- lm-jv--- lm-jv--- lmmjv--- --m----- lm-jv--- lmmjv--- DOLE Gare Routière 12:25 12:25 17:05 18:00 18:00 17:30 17:30 12:05 12:05 DOLE Place Grévy/Halte -

Janvier 2020
Bulletin Municipal JANVIER 2020 Parcey www.parcey.fr La Mairie vous informe Horaires Secrétariat Lundi 8h30 - 12h00 13h30 - 18h00 Mardi 8h30 - 12h00 13h30 - 17h30 Horaires Agence Postale Mercredi 8h30 - 12h00 FERMÉ Lundi FERMÉ 15h00 - 18h00 Jeudi 8h30 - 12h00 13h30 - 17h30 Mardi 9h00 - 12h00 FERMÉ Vendredi 8h30 - 12h00 FERMÉ Mercredi 9h00 - 12h00 FERMÉ Jeudi 9h00 - 12h00 FERMÉ Vendredi 9h00 - 12h00 FERMÉ CONTACT Mairie de PARCEY PERMANENCE DU DÉPUTÉ 1 rue d Aval M. Jean Marie SERMIER Mr Jacques FOURNIER, Mr Vincent DUGARDIN 39100 PARCEY 2ème jeudi de chaque mois en Mairie Mlle Lucie DEMONTROND Tél : 03.84.71.01.89 à partir de 14 H Mme Jennifer DAMY, Mme Sandrine BERTRAND Fax : 03.84.71.00.21 INFOS Elections municipales Date limite d’Inscription sur la liste électorale : 7 février 2020 En mairie ou sur service-public.fr Pièces à fournir : Carte nationale d’identité et justificatif de domicile : 01/2020 Responsable de la publication : J.P FAIVRE Maire Coordination de la rédaction : Commission Communication Crédits photos : Mairie - Dépôt légal à parution en mairie Document imprimé en Franche Comté - Conception et réalisation : Commission Communication CF www.parcey.fr La Mairie Chers concitoyens, chers amis, vous informe Notre village est fait d’un savant équilibre entre passé présent et avenir, entre équipement et projets, entre respon- sabilité, confiance et partage, le tout avec l’humain au cœur Horaires Secrétariat du village, pour garantir ce lien de proximité témoin du lieu Lundi 8h30 - 12h00 13h30 - 18h00 social et du « VIVRE ENSEMBLE -

Principaux Résultats Floristiques Des Prospections Des Cours D'eau Et Des
Les Nouvelles Archives de la Flore jurassienne, 5, 2007 – Société Botanique de Franche-Comté Principaux résultats floristiques des prospections des cours d’eau et des zones humides des vallées du Doubs et de la Loue par Marc Vuillemenot M. Vuillemenot, Conservatoire Botanique National de Franche-Comté, Porte Rivotte, 25000 Besançon. Courriel : [email protected] Résumé – La prospection de 340 kilomètres de rivières et d’annexes hydrauliques sur la vallée du Doubs et quelques-uns de ses affluents, destinée principalement à établir la typologie phytosociologique des végétations aquatiques, amphibies et rivulaires, a permis simultanément de réaliser de nombreuses obser- vations botaniques. Le présent article aborde ainsi la distribution et l’écologie de 80 taxons répertoriés sur ce territoire, dont 24 taxons de la liste rouge de Franche-Comté et 28 taxons considérés comme inva- sifs ou potentiellement invasifs dans la région. Mots-clés : inventaire floristique, plantes patrimoniales, plantes invasives, Doubs, Loue. Introduction taines observations botaniques réa- versant de 7 550 kilomètres carrés. lisées durant cette étude. L’essentiel Quatre grandes unités morpholo- et article reprend les prin- des descriptions est extrait du rap- giques sont classiquement définies cipaux résultats floristiques port de synthèse (Vuillemenot et de l’amont vers l’aval (Citterio, d’une étude réalisée en Hans, 2007), mais certaines ont Rollet et Piégay, 2003) : C2005 et 2006 par le Conservatoire pu être complétées par de nouvel- Botanique National de Franche- les données de la base Taxa©sbfc/ – le Doubs supérieur, ou Doubs de Comté (CBNFC), dans le cadre du cbfc et 24 taxons ont été nouvel- la Haute Chaîne, des sources à la programme « Avenir du Territoire lement commentés. -

DOLE NORD Liste D'ouvertures Fermetures Des Écoles-1.Xlsx
Direction des services départementaux de l'éducation nationale du Jura Mouvement social du 5 décembre. Nom de l'école Commune ouverte fermée George Sand Dole X St Exupéry Dole X Sorbiers élémentaire Dole X Sorbiers maternelle Dole X Rochebelle élémtaire Dole x Wilson Dole X Pointelin Dole X Rochebelle maternelle Dole X Beauregard maternelle X Beauregard élémentaire Dole X Rockefeller maternelle Dole X La Bedugue élémentaire Dole X La Bedugue maternelle Dole X Le Poiset élémentaire Dole X Le Poiset maternelle Dole x Amange Amange X Archelange Archelange X Authume Authume X Biarne Biarne X Chätenois Châtenois X Chevigny Chevigny X Foucherans maternelle Foucherans X Foucherans élémentaire Foucherans X Grédisans Grédisans X Jouhe Jouhe X Menotey Menotey X Moissey Moissey X Baverans Baverans X Brevans Brevans X Dammartin-Marpain Dammartin-Marpain X Montmirey-La-Ville Montmirey-La-Ville X Parcey Parcey X Rochefort-Sur-Nenon X Romange Romange X Villette-Les-Dole Villette-Les-Dole X Goux Goux x Ougney Ougney X Pagney Pagney X Vitreux Vitreux X Nom de l'école Commune ouverte fermée Abergement x Aiglepierre x Belmont X Chamblay X Champvans X Chissey X Choisey X Cramans X Damparis mat X Damparis elem X Dampierre X Dournon X Etrepigney X Evans X Fraisans elem X Fraisans mat X Gendrey X La Loye X La Vieille Loye X Marnoz X Mont/Vaudrey X Montbarrey X Mouchard X Nevy les Dole X Orchamps X Ounans X Port Lesney X Ranchot X Saint Aubin el X Saint Aubin mat X Salins Olivet X Salins Voltaire X Salins Chantemerle X Sampans X Santans X Sermange X Souvans X Tavaux J. -

CAPD Mouvement 14 Juin 2011
CAPD mouvement 14 juin 2011 Postes adaptés : 1 demande pour la rentrée 2011 (poste au CDDP de Lons) Bénéficiaires de l’obligation d’emploi (BOE) : Rentrée 2010 : 8 candidatures, 3 candidats se sont présentés devant le Jury et une candidature retenue. Rentrée 2011 : 5 candidatures, 3 candidats se sont présentés devant le Jury et une candidature retenue. Demandes de détachement / disponibilité / réintégration : Détachement : 10 demandes pour la rentrée 2011 Disponibilité : 8 demandes pour la rentrée 2011 Réintégration : 5 demandes pour la rentrée 2011 Renouvellement de disponibilité : 14 demandes pour la rentrée 2011 Couplages de temps partiels : 2010 : 173 demandes : 116 à 75% et 57 à 50% 2011 : 178 demandes : 123 à 75% et 55 à 50% 5 ETP se libèrent en plus cette année. Demandes d’inéat / exéat : Demandes des collègues titulaires : 6 demandes d’exéat et 25 demandes d’inéat pour le Jura. L’Inspecteur accorde les 6 exéats. La DRH attend les réponses des autres académies, mais déjà de nombreux refus d’exéats sont à signaler. A cette date, 3 collègues arriveraient par exéat dans le département du Jura. Demandes des collègues stagiaires : 15 demandes d’exéat et 6 demandes d’inéat pour le Jura. 3 accords de principe pour des échanges Doubs-Jura Avancement à la hors classe des PE au 1er septembre 2011 : Contingent de 19 promotions: FAIVRE OLIVER – BONNET PATRICE – CALLIGARIS PIERRE – GAUVIN BRIGITTE – TABERLET VERONIQUE – COILLOT ANNIE – MARCHAND NADINE – RIGAUD HERVE – ARBEZ CLAUDE – COMTE MARYVONNE – BEGUIN JEAN LUC – SIMON CHANTAL – MARCHIONINI PIERRE – PESENTI BRUNO – GRIFFON JOSETTE – NEYRAUD PAULE – DAUBIGNEY FRANCOIS – DESPRES MARIE LAURENCE – SCHERER CHANTAL Liste complémentaire : NICEY CHRISTIAN Equilibre poste-personne : Fin mai 2011 : 7 surnombres Prévisions rentrée 2011 : entre -15 et +5 (selon les inéats/exéats) en comptant les 18 stagiaires. -

L'homme Et La Loue
Rando-fiche - N°4 Chamblay Latitude : 46.9975133 Longitude : 5.709626 Niveau facile 4,3 km L’homme 6 m 2 h et la Loue PR La présence de l’eau, de la forêt, la diversité des paysages et Le Conseil la richesse du patrimoine naturel font du Val d’amour un lieu de l’OT ‘‘ Ce parcours agrémenté de 7 touristique remarquable. Axe de communication stratégique à tables de lecture lève le voile sur les l‘époque gallo-romaine, le Val d’amour est encore fortement relations, parfois conflictuelles et souvent complexes qui unissaient marqué par l’empreinte des activités du passé : Flottage des les habitants à la Loue’’ bois, anciens moulins, forêt de Chaux. Nous vous invitons à découvrir la rivière, le long d’un sentier au départ de Chamblay. Le paysage conserve en mémoire les traces des divagations de la Loue et révèle comment l’Homme, a exploité les ressources de la rivière et lutté contre les crues. L E S www.tourisme-paysdedole.fr P E C T A C L E E N C O N T I N U Dole Forêt de Chaux Lons/Genève Doubs D 405 La loue Parcey Chamblay MouchardL’homme / Salins et la Loue La loue D 472 Ounans Souvans Vaudrey Lons ACCES Depuis l’office de tourisme, place Grévy, suivre direction Lons-le-Saunier/Genève, traverser Parcey. Dole À la sortie de Parcey, suivre Poligny. Forêt de Chaux Environ 1 km après avoir traversé Souvans, au rond-point, prendre la dernière sortie Lons/Genève direction Mont-sous-Vaudrey/Arbois. -

Janvier 2016
Bulletin Municipal JANVIER 2016 Parcey vie Municipale La Mairie vous informe Horaires Secrétariat CONTACT Mairie de PARCEY 1 rue d Aval 39100 PARCEY Tél : 03.84.71.01.89 Fax : 03.84.71.00.21 : Taille des Arbres Vous devez veiller à ce que vos propres plantations n'empiètent pas sur le domaine public : vos arbres INFOS ou vos haies ne doivent pas empêcher de marcher sur un trottoir ou constituer un danger pour la circulation routière. Si tel est le cas, le maire peut BACS jaunes et bleus vous contraindre à élaguer vos arbres en vous adressant une injonction de faire. L'article L2212-2-2 Ramassage des bacs jaunes et bleus les du Code général des collectivités territoriales prévoit qu'en cas de mise en demeure sans résultat, le maire peut procéder à l'exécution semaines paires à compter de forcée des travaux d'élagage, les frais janvier 2016 afférents aux opérations étant alors mis à la charge des propriétaires négligents. Notre village bouge et s’agrandit Bienvenue aux nouveaux commerçants et habitants Bulletin - 01/2016 Responsable de la publication : J.P FAIVRE Maire - Coordination de la rédaction : Commission communication Crédits photos : Mairie - Dépôt légal à parution en mairie Document imprimé en Franche Comté - Conception et réalisation : Commission Communication CF vie Cela fait maintenant plus d’un an que la nouvelle équipe municipale est au travail et développe de nouveaux projets, tout en assurant une gestion rigoureuse des budgets. Dans un contexte où l’Etat prévoit une baisse des dotations Municipale accordées aux communes, des choix sont à faire sur les investissements et aussi sur la gestion courante. -

Mouchard Du Bois, Des Trains Et Beaucoup De Vie… P
N°211 Janvier 2019 DOLE 53, avenue Eisenhower 03 84 72 34 34 Pays Dolois [email protected] Journal GRATUIT d’informations Fondu de Jura GROS OEUVRE MENUISERIE Crémant du Jura CARRELAGE Marcel Cabelier €99 75 cl SANITAIRE Zone Commerciale des Épenottes - Dole 5 OUTILLAGE L’abus d’alcool est dangereux pour la santé à consommer avec modération Jusqu’au 20 février 2019 - [email protected] - 39500 TAVAUX Z.I. “Les Charmes” - 03 84 81 97 03 - [email protected] Mouchard Du bois, des trains et beaucoup de vie… p. 20 DOLE - REPONSE NATURE Tourisme 65 av. Eisenhower - 03 84 82 11 29 MarieThé Brocard, une Pages élue qui s’investit pour le spéciales développement… p. 4 Pages 15 à 19 70 ans de Scrabble à Dole, Parcey, Annoire p. 12 Conservatoire, copistes, lecture, cinéma, idées… à chacun sa nuit ! p. 26 Venez les découvrir ! PORTES OUVERTES 18 - 19 - 20 JANVIER Nous vous souhaitons une très bonne année 2019 ! L’équipe Automobiles Cuynet AUTOMOBILES CUYNET - Zone commerciale de Choisey - 39100 DOLE - 03 84 82 21 11 - www.cuynet.fr volkswagen.fr 2 actus Pays Dolois n°211 - janvier 2019 Santé 1 Un bol d’air pour l’hôpital de Dole ? Le centre hospitalier de Dole a signé une convention avec le centre hospitalier universitaire de Besançon. L’objectif ? Développer une offre de chirurgie ambulatoire de proximité, adaptée aux besoins de la population. Des consultations dans de nouvelles disciplines vont être proposées dès ce début d’année et une rénovation des blocs chirurgicaux est prévue pour 2020. Le centre hospitalier Louis Pasteur de Dole pour- priorité pour l’hôpital de Dole de s’assurer d’un suit la coopération avec le centre hospitalier régio- maximum de soins” souligne le maire de la Ville, nal et universitaire Jean Minjoz. -

Madame De Chamblay
Alexandre Dumas MadameMadame dede ChamblayChamblay BeQ Alexandre Dumas Madame de Chamblay roman La Bibliothèque électronique du Québec Collection À tous les vents Volume 1350 : version 1.0 2 Du même auteur, à la Bibliothèque : Les Louves de Machecoul Les mille et un fantômes La femme au collier de velours Les mariages du père Olifus Le prince des voleurs Robin Hood, le proscrit Les compagnons de Jéhu Le comte de Monte-Cristo La San Felice La reine Margot Les trois mousquetaires Le vicomte de Bragelonne Le chevalier de Maison-Rouge Histoire d’un casse noisette et autres contes La bouillie de la comtesse Berthe et autres contes 3 Madame de Chamblay Édition de référence : Paris, Michel Lévy Frères, 1865. Numérisation : Bibliothèque numérique romande. Relecture : Jean-Yves Dupuis. 4 Quelques mots au lecteur C’est une singulière histoire que celle que je vais vous raconter – ou plutôt que celle que l’on va vous raconter, cher lecteur. Elle est écrite par un homme qui n’a jamais rien écrit que cette histoire. C’est une page détachée de sa vie, ou, pour mieux dire, c’est sa vie tout entière. La vie de l’homme se mesure, non point par le nombre d’années pendant lesquelles il a existé, mais par les minutes pendant lesquelles son cœur a battu. Tel vieillard, mort à quatre-vingts ans, n’a vécu parfois en réalité qu’un an, qu’un mois, qu’un jour. Vivre, c’est être heureux ou souffrir. Faites passer devant le moribond couché sur son lit d’agonie tous les jours qu’il a traversés, il 5 ne reconnaîtra que ceux qui viendront à lui le rire sur les lèvres ou les larmes dans les yeux. -

Ligne Dole/Arbois AR
Période scolaire Période vacances ARBOIS Lundi, mardi, DOLE ARBOIS jeudi, Mercredi Lundi, mardi, jeudi, vendredi Mercredi Lundi, mardi, jeudi, vendredi Lundi à vendredi VADANS vendredi ARBOIS DOLE MATHENAY MOLAMBOZ 12:05 17:30 12:05 17:30 DOLE Gare Routière 12:25 12:25 17:05 18:05 18:05 LA FERTE DOLE Avenue de Lahr 12:09 12:29 12:29 16:30 17:10 18:09 18:09 12:09 HORAIRES VALABLES À PARTIR DU 7 OCTOBRE 2019 VAUDREY DOLE CES Ledoux 12:10 16:40 MONT-SOUS-VAUDREY DOLE-GOUX Mairie 12:14 12:37 16:44 18:17 DOLE-GOUX Place de la Cornée 12:15 12:39 16:45 18:19 LR 312 BANS LA LOYE Place 12:20 12:43 16:50 18:21 SOUVANS AUGERANS Place 12:25 12:46 16:56 18:26 NEVY-LES-DOLE LA VIEILLE-LOYE Mairie 12:51 18:31 MOUCHARD OUNANS Eglise 13:00 18:38 VILLETTE-LES-DOLE Café Maublanc 17:20 VILLERS-FARLAY NEVY-LES DOLE Place Bascule 17:27 ECLEUX SOUVANS Carrefour Gare 17:29 CHAMBLAY SOUVANS Poste 17:30 OUNANS SOUVANS RN 63 17:32 CHISSEY-SUR-LOUE MONT-SOUS-VAUDREY École 13:05 17:36 VAUDREY Église 13:09 17:41 18:42 CHATELAY VAUDREY Pont 13:10 17:43 18:43 GERMIGNEY LA FERTE Chalet 13:12 17:48 18:47 SANTANS MATHENAY Molamboz 13:14 17:50 18:49 MONTBARREY VADANS Carrefour RD 469 13:16 17:52 18:51 ARBOIS Arbois Tourisme 13:21 17:56 18:56 BELMONT ARBOIS La Foule 13:23 18:00 18:58 LA VIEILLE-LOYE BELMONT Rue du Val d'Amour 12:32 12:46 17:03 18:26 AUGERANS MONTBARREY Place 12:54 18:30 SANTANS Place 12:56 18:39 LA LOYE GERMIGNEY Monuments aux morts 12:58 18:43 emmène-moi là où je veux ! DOLE-GOUX CHATELAY Village