HMGN3 / TRIP7 (1-77, His-Tag) Human Protein Product Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Transcriptional Control of Tissue-Resident Memory T Cell Generation
Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2019 © 2019 Filip Cvetkovski All rights reserved ABSTRACT Transcriptional control of tissue-resident memory T cell generation Filip Cvetkovski Tissue-resident memory T cells (TRM) are a non-circulating subset of memory that are maintained at sites of pathogen entry and mediate optimal protection against reinfection. Lung TRM can be generated in response to respiratory infection or vaccination, however, the molecular pathways involved in CD4+TRM establishment have not been defined. Here, we performed transcriptional profiling of influenza-specific lung CD4+TRM following influenza infection to identify pathways implicated in CD4+TRM generation and homeostasis. Lung CD4+TRM displayed a unique transcriptional profile distinct from spleen memory, including up-regulation of a gene network induced by the transcription factor IRF4, a known regulator of effector T cell differentiation. In addition, the gene expression profile of lung CD4+TRM was enriched in gene sets previously described in tissue-resident regulatory T cells. Up-regulation of immunomodulatory molecules such as CTLA-4, PD-1, and ICOS, suggested a potential regulatory role for CD4+TRM in tissues. Using loss-of-function genetic experiments in mice, we demonstrate that IRF4 is required for the generation of lung-localized pathogen-specific effector CD4+T cells during acute influenza infection. Influenza-specific IRF4−/− T cells failed to fully express CD44, and maintained high levels of CD62L compared to wild type, suggesting a defect in complete differentiation into lung-tropic effector T cells. -

Hepatitis C Virus As a Unique Human Model Disease to Define
viruses Review Hepatitis C Virus as a Unique Human Model Disease to Define Differences in the Transcriptional Landscape of T Cells in Acute versus Chronic Infection David Wolski and Georg M. Lauer * Liver Center at the Gastrointestinal Unit, Department of Medicine, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA * Correspondence: [email protected]; Tel.: +1-617-724-7515 Received: 27 June 2019; Accepted: 23 July 2019; Published: 26 July 2019 Abstract: The hepatitis C virus is unique among chronic viral infections in that an acute outcome with complete viral elimination is observed in a minority of infected patients. This unique feature allows direct comparison of successful immune responses with those that fail in the setting of the same human infection. Here we review how this scenario can be used to achieve better understanding of transcriptional regulation of T-cell differentiation. Specifically, we discuss results from a study comparing transcriptional profiles of hepatitis C virus (HCV)-specific CD8 T-cells during early HCV infection between patients that do and do not control and eliminate HCV. Identification of early gene expression differences in key T-cell differentiation molecules as well as clearly distinct transcriptional networks related to cell metabolism and nucleosomal regulation reveal novel insights into the development of exhausted and memory T-cells. With additional transcriptional studies of HCV-specific CD4 and CD8 T-cells in different stages of infection currently underway, we expect HCV infection to become a valuable model disease to study human immunity to viruses. Keywords: viral hepatitis; hepatitis C virus; T cells; transcriptional regulation; transcription factors; metabolism; nucleosome 1. -

Virtual Chip-Seq: Predicting Transcription Factor Binding
bioRxiv preprint doi: https://doi.org/10.1101/168419; this version posted March 12, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Virtual ChIP-seq: predicting transcription factor binding 2 by learning from the transcriptome 1,2,3 1,2,3,4,5 3 Mehran Karimzadeh and Michael M. Hoffman 1 4 Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada 2 5 Princess Margaret Cancer Centre, Toronto, ON, Canada 3 6 Vector Institute, Toronto, ON, Canada 4 7 Department of Computer Science, University of Toronto, Toronto, ON, Canada 5 8 Lead contact: michael.hoff[email protected] 9 March 8, 2019 10 Abstract 11 Motivation: 12 Identifying transcription factor binding sites is the first step in pinpointing non-coding mutations 13 that disrupt the regulatory function of transcription factors and promote disease. ChIP-seq is 14 the most common method for identifying binding sites, but performing it on patient samples is 15 hampered by the amount of available biological material and the cost of the experiment. Existing 16 methods for computational prediction of regulatory elements primarily predict binding in genomic 17 regions with sequence similarity to known transcription factor sequence preferences. This has limited 18 efficacy since most binding sites do not resemble known transcription factor sequence motifs, and 19 many transcription factors are not even sequence-specific. 20 Results: 21 We developed Virtual ChIP-seq, which predicts binding of individual transcription factors in new 22 cell types using an artificial neural network that integrates ChIP-seq results from other cell types 23 and chromatin accessibility data in the new cell type. -

Encoded on Chromosome 6P21.33 in Human Breast Cancers Revealed by Transcrip- Tome Analysis Yan A
Journal of Cancer 2010, 1 38 Journal of Cancer 2010; 1:38-50 © Ivyspring International Publisher. All rights reserved Research Paper Undetectable and Decreased Expression of KIAA1949 (Phostensin) Encoded on Chromosome 6p21.33 in Human Breast Cancers Revealed by Transcrip- tome Analysis Yan A. Su1 , Jun Yang1, Lian Tao1, and Hein Nguyen1, and Ping He2 1. GenProMarkers Inc., Rockville, Maryland 20850, USA; 2. Division of Hematology, Center for Biological Evaluation and Research, Food and Drug Administration, Bethesda, MD 20892, USA Corresponding author: Yan A. Su, MD, PhD, GenProMarkers Inc., 9700 Great Seneca Highway, Suite 182, Rockville, Maryland 20850. Phone: (301) 326-6523; FAX: (240) 453-6208; Email:[email protected] Published: 2010.06.21 Abstract Cytogenetic aberration and loss of heterozygosity (LOH) are documented on chromosome 6 in many cancers and the introduction of a neo-tagged chromosome 6 into breast cancer cell lines mediates suppression of tumorigenicity. In this study, we described the identification of KIAA1949 (phostensin) as a putative tumor suppressor gene. Our microarray analysis screened 25,985 cDNAs between a tumorigenic and metastatic breast cancer cell line MDA-MB-231 and the chromosome 6-mediated suppressed, non-tumorigenic and non-metastatic derivative cell line MDA/H6, resulting in the identification of 651 differentially expressed genes. Using customized microarrays containing these 651 cDNAs and 117 con- trols, we identified 200 frequently dysregulated genes in 10 breast cancer cell lines and 5 tumor tissues using MDA/H6 as reference. Our bioinformatics analysis revealed that chro- mosome 6 encodes 25 of these 200 genes, with 4 downregulation and 21 upergulation. -

Identification of Genomic Targets of Krüppel-Like Factor 9 in Mouse Hippocampal
Identification of Genomic Targets of Krüppel-like Factor 9 in Mouse Hippocampal Neurons: Evidence for a role in modulating peripheral circadian clocks by Joseph R. Knoedler A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Neuroscience) in the University of Michigan 2016 Doctoral Committee: Professor Robert J. Denver, Chair Professor Daniel Goldman Professor Diane Robins Professor Audrey Seasholtz Associate Professor Bing Ye ©Joseph R. Knoedler All Rights Reserved 2016 To my parents, who never once questioned my decision to become the other kind of doctor, And to Lucy, who has pushed me to be a better person from day one. ii Acknowledgements I have a huge number of people to thank for having made it to this point, so in no particular order: -I would like to thank my adviser, Dr. Robert J. Denver, for his guidance, encouragement, and patience over the last seven years; his mentorship has been indispensable for my growth as a scientist -I would also like to thank my committee members, Drs. Audrey Seasholtz, Dan Goldman, Diane Robins and Bing Ye, for their constructive feedback and their willingness to meet in a frequently cold, windowless room across campus from where they work -I am hugely indebted to Pia Bagamasbad and Yasuhiro Kyono for teaching me almost everything I know about molecular biology and bioinformatics, and to Arasakumar Subramani for his tireless work during the home stretch to my dissertation -I am grateful for the Neuroscience Program leadership and staff, in particular -

Human Papillomavirus Oncoprotein E7 Dysregulates Immune
HUMAN PAPILLOMAVIRUS ONCOPROTEIN E7 DYSREGULATES IMMUNE RESPONSES THROUGH EPIGENETIC MANIPULATION by LOUIS J CICCHINI B.S., University of Arizona, 2009 A thesis submitted to the Faculty of the Graduate School of the University of Colorado in partial fulfillment of the requirements for the degree of Doctor of Philosophy Molecular Biology 2016 This thesis for the Doctor of Philosophy degree by Louis J Cicchini has been approved for the Molecular Biology Program by Rytis Prekeris, Chair Dohun Pyeon, Advisor James Hagman Thomas E. Morrison Xiao-Jing Wang Date: August 18, 2016 ii Cicchini, Louis J. (PhD, Molecular Biology) Human Papillomavirus Oncoprotein E7 Dysregulates Immune Responses through Epigenetic Manipulation Thesis directed by Associate Professor Dohun Pyeon ABSTRACT High-risk human papillomaviruses (HPVs) are causally associated with multiple human cancers. Previous studies have shown that the HPV oncoprotein E7 induces immune suppression; however, the underlying mechanisms remain unknown. We report that, while expression of many proinflammatory chemokines increases throughout HPV-positive cancer progression, CXCL14 is dramatically downregulated by promoter hypermethylation in an E7- dependent manner. Our in vivo mouse models revealed that restoration of Cxcl14 expression in HPV-positive mouse oropharyngeal carcinoma cells clears tumors in immunocompetent syngeneic mice, but not in Rag1-deficient mice. Further, restoration of Cxcl14 expression significantly increases natural killer (NK), CD4+ T, and CD8+ T cell infiltration into the tumor-draining lymph nodes in vivo. In vitro transwell migration assays show that restoration of Cxcl14 expression induces chemotaxis of NK, CD4+ T, and CD8+ T cells. These findings suggest that high-risk HPV E7 is likely to dysregulate host gene expression in order to persist by modulating host DNA methylation. -

Supplemental Figure 1. Protein-Protein Interaction Network with Increased Expression in Fteb During the Luteal Phase
Supplemental Figure 1. Protein-protein interaction network with increased expression in FTEb during the luteal phase. Supplemental Figure 2. Protein-protein interaction network with decreased expression in FTEb during luteal phase. LEGENDS TO SUPPLEMENTAL FIGURES Supplemental Figure 1. Protein-protein interaction network with increased expression in FTEb during the luteal phase. Submission of probe sets differentially expressed in the FTEb specimens that clustered with SerCa as well as those specifically altered in FTEb luteal samples to the online I2D database revealed overlapping networks of proteins with increased expression in the four FTEb samples and/or FTEb luteal samples overall. Proteins are represented by nodes, and known and predicted first-degree interactions are represented by solid lines. Genes encoding proteins shown as large ovals highlighted in blue were exclusively found in the first comparison (Manuscript Figure 2), whereas those highlighted in red were only found in the second comparison (Manuscript Figure 3). Genes encoding proteins shown as large ovals highlighted in black were found in both comparisons. The color of each node indicates the ontology of the corresponding protein as determined by the Online Predicted Human Interaction Database (OPHID) link with the NAViGaTOR software. Supplemental Figure 2. Protein-protein interaction network with decreased expression in FTEb during the luteal phase. Submission of probe sets differentially expressed in the FTEb specimens that clustered with SerCa as well as those specifically altered in FTEb luteal samples to the online I2D database revealed overlapping networks of proteins with decreased expression in the four FTEb samples and/or FTEb luteal samples overall. Proteins are represented by nodes, and known and predicted first-degree interactions are represented by solid lines. -

Knockdown of High Mobility Group Box 3 Impairs Cell Viability and Colony Formation but Increases Apoptosis in A549 Human Non‑Small Cell Lung Cancer Cells
ONCOLOGY LETTERS 17: 2937-2945, 2019 Knockdown of high mobility group box 3 impairs cell viability and colony formation but increases apoptosis in A549 human non‑small cell lung cancer cells NING SONG1*, BAOHUA WANG2*, GUISHAN FENG3, LIN DUAN1, SHENGFANG YUAN1, WEIHUA JIA1 and YI LIU1 Departments of 1Infectious Diseases and 2Thoracic Surgery, The Second Hospital of Hebei Medical University, Shijiazhuang, Hebei 050000; 3Department of Internal Medicine, Hebei Yi County Hospital, Baoding, Hebei 074200, P.R. China Received August 25, 2017; Accepted February 22, 2018 DOI: 10.3892/ol.2019.9927 Abstract. Previous research has linked high mobility group of NSCLC; however, owing to the heterogeneity and genetic box 3 (HMGB3) overexpression to the malignant progression instability of NSCLC cells, therapeutic methods that are being and poor prognosis of non-small cell lung cancer (NSCLC). The used in the clinic, including chemotherapy, radiation therapy present study investigated the role of HMGB3 in cell survival or targeted therapy such as epithelial growth factor receptor and colony formation of NSCLC cells. Stable knockdown of (EGFR)-tyrosine kinase inhibitors, often exhibit short-lived HMGB3 in A549 cells was achieved by lentiviral-based shRNA treatment response (4). Novel therapeutic targets or methods interference and verified by detection of the transcriptional and are therefore required. translational level of HMGB3 with reverse transcription-quan- A previous study revealed that high mobility group box 3 titative polymerase chain reaction and western blotting, (HMGB3) overexpression is an independent risk factor for respectively. The influence of HMGB3 knockdown on A549 NSCLC progression, lymph node metastasis and poor survival cell viability and apoptotic rate was evaluated by Cell Counting rates of patients (5), indicating the prognostic value and the onco- Kit‑8 assay and flow cytometry following annexin V staining, genic role of HMGB3 overexpression in NSCLC development. -

Systematic Discovery and Characterization of Regulatory Motifs in ENCODE TF Binding Experiments Pouya Kheradpour1,2 and Manolis Kellis1,2,*
Nucleic Acids Research Advance Access published December 13, 2013 Nucleic Acids Research, 2013, 1–12 doi:10.1093/nar/gkt1249 Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments Pouya Kheradpour1,2 and Manolis Kellis1,2,* 1Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, 32 Vassar St, Cambridge, MA 02139, USA and 2Broad Institute of MIT and Harvard, 7 Cambridge Center, Cambridge, MA 02139, USA Received August 7, 2013; Revised November 6, 2013; Accepted November 7, 2013 Downloaded from ABSTRACT present in a given condition and cell type or tissue. As these technologies have matured, their use has become Recent advances in technology have led to a increasingly widespread. The resolution of these experi- dramatic increase in the number of available tran- mental techniques can be as low as 300 bp for ChIP-chip scription factor ChIP-seq and ChIP-chip data sets. (5) and 50 bp for ChIP-seq (6), depending on the experi- Understanding the motif content of these data sets mental design (e.g. fragment size, paired-end sequencing) http://nar.oxfordjournals.org/ is an important step in understanding the underlying and algorithmic processing of the raw data. mechanisms of regulation. Here we provide a sys- The use of these technologies on a variety of factors tematic motif analysis for 427 human ChIP-seq data across many cell types has increasingly highlighted the sets using motifs curated from the literature and complex nature of TF activity, often violating the simple also discovered de novo using five established model of a factor binding to its recognition pattern (motif) motif discovery tools. -

Supplemental Solier
Supplementary Figure 1. Importance of Exon numbers for transcript downregulation by CPT Numbers of down-regulated genes for four groups of comparable size genes, differing only by the number of exons. Supplementary Figure 2. CPT up-regulates the p53 signaling pathway genes A, List of the GO categories for the up-regulated genes in CPT-treated HCT116 cells (p<0.05). In bold: GO category also present for the genes that are up-regulated in CPT- treated MCF7 cells. B, List of the up-regulated genes in both CPT-treated HCT116 cells and CPT-treated MCF7 cells (CPT 4 h). C, RT-PCR showing the effect of CPT on JUN and H2AFJ transcripts. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. Supplementary Figure 3. Down-regulation of RNA degradation-related genes after CPT treatment A, “RNA degradation” pathway from KEGG. The genes with “red stars” were down- regulated genes after CPT treatment. B, Affy Exon array data for the “CNOT” genes. The log2 difference for the “CNOT” genes expression depending on CPT treatment was normalized to the untreated controls. C, RT-PCR showing the effect of CPT on “CNOT” genes down-regulation. HCT116 cells were treated with CPT (10 µM, 20 h) and CNOT6L, CNOT2, CNOT4 and CNOT6 mRNA were analysed by RT-PCR. Control cells were exposed to DMSO. β2 microglobulin (β2) mRNA was used as control. D, CNOT6L down-regulation after CPT treatment. CNOT6L transcript was analysed by Q- PCR. Supplementary Figure 4. Down-regulation of ubiquitin-related genes after CPT treatment A, “Ubiquitin-mediated proteolysis” pathway from KEGG. -
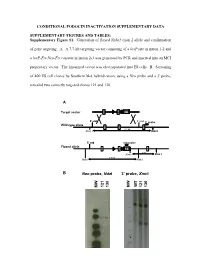
Supplementary Figure S1. Generation of Floxed Nphs2 Exon 2 Allele and Confirmation
CONDITIONAL PODOCIN INACTIVATION SUPPLEMENTARY DATA SUPPLEMENTARY FIGURES AND TABLES: Supplementary Figure S1. Generation of floxed Nphs2 exon 2 allele and confirmation of gene targeting. A. A 7.7-kb targeting vector consisting of a loxP site in intron 1-2 and a loxP-Frt-Neo-Frt cassette in intron 2-3 was generated by PCR and inserted into an MCI proprietary vector. The linearized vector was electroporated into ES cells. B. Screening of 400 ES cell clones by Southern blot hybridization, using a Neo probe and a 3' probe, revealed two correctly targeted clones 121 and 130. A Target vector 1 2 NEO 5’ end 3’ end 3’ probe Wild-type allele 1 2 3 9.2 kb Xmn I Xmn I 5’ end neo probe Floxed allele 1 2 NEO 3 5 kb Xmn I Xmn I 7.8 kb Nde I Nde I B Neo probe, NdeI 3’ probe, XmnI WT 121 130 121 130 MW MW - 1 - Supplementary Figure S2. Generation of Nphs2lox2/-,Cre+ mice and excision of exon 2 upon Cre recombinase induction. A. Triallelic Nphs2lox2/-,Cre+ mice were obtained by mating phenotypically normal Nphs2lox2/lox2 mice with Nphs2+/-,Cre+ mice. Mendelian inheritance of these alleles was observed. B. Genotypes were verified by multiplex PCR of tail genomic DNA. C. Cre recombinase activity was induced upon tamoxifen administration, leading to excision of the floxed exon 2 of the Nphs2 gene. D. Cre recombinase activity in the kidney was verified by PCR using a set of forward and reverse primers designed around exon 2 and demonstrating a 692-bp product (before Cre) and a 316-bp product (after Cre) using genomic DNA extracted from the renal cortex.