Psychostimulant-Regulated Plasticity in Interneurons of the Nucleus Accumbens
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genetic Analysis of the Calcineurin Pathway Identifies Members of the EGR Gene Family, Specifically EGR3, As Potential Susceptibility Candidates in Schizophrenia
Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia Kazuo Yamada*, David J. Gerber†, Yoshimi Iwayama*, Tetsuo Ohnishi*, Hisako Ohba*, Tomoko Toyota*, Jun Aruga‡, Yoshio Minabe*§, Susumu Tonegawa†¶, and Takeo Yoshikawa*ʈ** Laboratories for *Molecular Psychiatry and ‡Comparative Neural Development, RIKEN Brain Science Institute, Saitama 351-0198, Japan; †Howard Hughes Medical Institute and RIKEN–MIT Neuroscience Research Center, The Picower Institute for Learning and Memory, Massachusetts Institute of Technology, Cambridge, MA 02139; §Department of Psychiatry and Neurology, Kanazawa University School of Medicine, Ishikawa 920-8641, Japan; and ʈCore Research for Evolutional Science and Technology, Japan Science and Technology Agency, Saitama 332-0012, Japan Contributed by Susumu Tonegawa, December 7, 2006 (sent for review September 22, 2006) The calcineurin cascade is central to neuronal signal transduction, cineurin is particularly enriched in the CNS, where it plays a critical and genes in this network are intriguing candidate schizophrenia role in the regulation of a diverse array of neuronal functions (5, 6). susceptibility genes. To replicate and extend our previously re- Interestingly, calcineurin is positioned downstream of dopaminer- ported association between the PPP3CC gene, encoding the cal- gic signaling (7) and is involved in NMDA receptor-mediated cineurin catalytic ␥-subunit, and schizophrenia, we examined 84 synaptic plasticity (8) and could therefore provide an important SNPs from 14 calcineurin-related candidate genes for genetic as- functional link between these two neurotransmitter systems. To sociation by using 124 Japanese schizophrenic pedigrees. Four of further explore the involvement of calcineurin dysfunction in these genes (PPP3CC, EGR2, EGR3, and EGR4) showed nominally schizophrenia, we have tested for genetic association of a subset of significant association with schizophrenia. -

Egr-1 Induces DARPP-32 Expression in Striatal Medium Spiny Neurons Via a Conserved Intragenic Element
6808 • The Journal of Neuroscience, May 16, 2012 • 32(20):6808–6818 Cellular/Molecular Egr-1 Induces DARPP-32 Expression in Striatal Medium Spiny Neurons via a Conserved Intragenic Element Serene Keilani,1 Samira Chandwani,1 Georgia Dolios,2 Alexey Bogush,3 Heike Beck,4 Antonis K. Hatzopoulos,5 Gadiparthi N. Rao,6 Elizabeth A. Thomas,7 Rong Wang,2 and Michelle E. Ehrlich1,2 Departments of 1Neurology and Pediatrics and 2Genetics and Genomic Sciences, Mount Sinai School of Medicine, New York, New York 10029, 3Weinberg Unit for ALS Research, Department of Neuroscience, Farber Institute for Neuroscience, Thomas Jefferson University, Philadelphia, Pennsylvania 19107, 4Walter Brendel Center of Experimental Medicine, Ludwig Maximilians University, D-81377 Munich, Germany, 5Division of Cardiovascular Medicine, Department of Medicine, Vanderbilt University, Nashville, Tennessee 37240, 6Department of Physiology, University of Tennessee Health Science Center, Memphis, Tennessee 38163, and 7Department of Molecular Biology, The Scripps Research Institute, La Jolla, California 92037 DARPP-32 (dopamine and adenosine 3Ј,5Ј-cyclic monophosphate cAMP-regulated phosphoprotein, 32 kDa) is a striatal-enriched protein that mediates signaling by dopamine and other first messengers in the medium spiny neurons. The transcriptional mechanisms that regulate striatal DARPP-32 expression remain enigmatic and are a subject of much interest in the efforts to induce a striatal phenotype in stem cells. We report the identification and characterization of a conserved region, also known as H10, in intron IV of the gene that codes for DARPP-32 (Ppp1r1b). This DNA sequence forms multiunit complexes with nuclear proteins from adult and embry- onic striata of mice and rats. -

DRD2 and PPP1R1B (DARPP-32)
Hettinger et al. Behavioral and Brain Functions 2012, 8:19 http://www.behavioralandbrainfunctions.com/8/1/19 RESEARCH Open Access DRD2 and PPP1R1B (DARPP-32) polymorphisms independently confer increased risk for autism spectrum disorders and additively predict affected status in male-only affected sib-pair families Joe A Hettinger1, Xudong Liu2,3, Melissa L Hudson2,3,4, Alana Lee2,3,4, Ira L Cohen4,5, Ron C Michaelis6, Charles E Schwartz7, Suzanne ME Lewis4,8,9 and Jeanette JA Holden1,2,3,4,10,11* Abstract Background: The neurotransmitter dopamine (DA) modulates executive functions, learning, and emotional processing, all of which are impaired in individuals with autism spectrum disorders (ASDs). Our previous findings suggest a role for dopamine-related genes in families with only affected males. Methods: We examined two additional genes which affect DA function, the DRD2 and PPP1R1B (DARPP-32) genes, in a cohort of 112 male-only affected sib-pair families. Selected polymorphisms spanning these genes were genotyped and both family-based and population-based tests were carried out for association analysis. General discriminant analysis was used to examine the gene-gene interactions in predicting autism susceptibility. Results: There was a significantly increased frequency of the DRD2 rs1800498TT genotype (P = 0.007) in affected males compared to the comparison group, apparently due to over-transmission of the T allele (P = 0.0003). The frequency of the PPP1R1B rs1495099CC genotype in affected males was also higher than that in the comparison group (P = 0.002) due to preferential transmission of the C allele from parents to affected children (P = 0.0009). -

Effects of PPP1R1B (DARPP-32) Polymorphism on Feedback-Related Brain Potentials Across the Life Span
ORIGINAL RESEARCH ARTICLE published: 04 March 2013 doi: 10.3389/fpsyg.2013.00089 Effects of PPP1R1B (DARPP-32) polymorphism on feedback-related brain potentials across the life span Dorothea Hämmerer 1,2*†, Gudio Biele 3,4,Viktor Müller 1, HolgerThiele 5, Peter Nürnberg 5,6,7, Hauke R. Heekeren1,3 and Shu-Chen Li 1,2*† 1 Center for Lifespan Psychology, Max Planck Institute for Human Development, Berlin, Germany 2 Department of Psychology, Technische Universität Dresden, Dresden, Germany 3 Department of Education and Psychology, Freie Universität Berlin, Berlin, Germany 4 Institute of Psychology, University of Oslo, Oslo, Norway 5 Cologne Center for Genomics, University of Cologne, Cologne, Germany 6 Center for Molecular Medicine Cologne, University of Cologne, Cologne, Germany 7 Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases, University of Cologne, Cologne, Germany Edited by: Maximizing gains during probabilistic reinforcement learning requires the updating of Alain Dagher, Montreal Neurological choice – outcome expectations at the time when the feedback about a specific choice Institute and Hospital, Canada or action is given. Extant theories and evidence suggest that dopaminergic modulation Reviewed by: Meghana Bhatt, City of Hope, USA plays a crucial role in reinforcement learning and the updating of choice – outcome expec- James F.Cavanagh, Brown University, tations. Furthermore, recently a positive component of the event-related potential about USA 200 ms (P2) after feedback has been suggested to reflect such updating. The efficacy of *Correspondence: dopaminergic modulation changes across the life span. However, to date investigations Dorothea Hämmerer and Shu-Chen of age-related differences in feedback-related P2 during reinforcement learning are still Li, Department of Psychology, Technische Universität Dresden, scarce. -
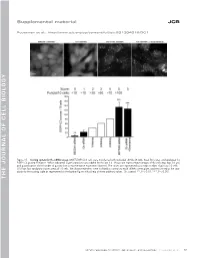
Thejournalofcellb Io Logy
Supplemental material JCB Puustinen et al., http://www.jcb.org/cgi/content/full/jcb.201304012/DC1 Figure S1. Scoring system for the siRNA screen. MCF7-EGFP-LC3 cells were transfected with indicated siRNAs (8 nM), fixed 56 h later, and analyzed for EGFP-LC3 puncta formation. When indicated, 5 µM siramesine was added for the last 3 h. Shown are representative images of the cells (top; bar, 10 µm) and quantification of the number of puncta from a representative experiment (bottom). The values are represented as a mean number of puncta/10 cells ± SDs from four randomly chosen areas of 10 cells. The shown treatments were included as controls to each siRNA screen plate, and they served as the stan- dards for the scoring scale as represented on the bottom figure with a help of three arbitrary values. Ctr, control. **, P < 0.01; ***, P < 0.001. THE JOURNAL OF CELL BIOLOGY CIP2A regulates mTORC1, cell growth, and autophagy • Puustinen et al. S1 Figure S2. A cartoon highlighting the positions of the candidate autophagy-regulating genes in the insulin receptor signaling network. The cartoon was prepared by Ingenuity Pathway Analysis. software. CP, carbohydrate phosphatase; ERK, extracellular signal–regulated kinase; FSH, follicle-stimulating hor- mone; EG, epidermal growth factor. S2 JCB Figure S3. The effect of siRNAs targeting the autophagy-regulating PP2 subunits on target gene expression, cell density, and mTORC1 pathway. (A) Total RNA was isolated from MCF7 cells treated with indicated siRNAs (20 nM) for 54 h and analyzed for target gene expression by qPCR using primers de- signed to specifically amplify the indicated PP2A-related mRNAs andACTB mRNA (internal control). -

CSE642 Final Version
Eindhoven University of Technology MASTER Dimensionality reduction of gene expression data Arts, S. Award date: 2018 Link to publication Disclaimer This document contains a student thesis (bachelor's or master's), as authored by a student at Eindhoven University of Technology. Student theses are made available in the TU/e repository upon obtaining the required degree. The grade received is not published on the document as presented in the repository. The required complexity or quality of research of student theses may vary by program, and the required minimum study period may vary in duration. General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain Eindhoven University of Technology MASTER THESIS Dimensionality Reduction of Gene Expression Data Author: S. (Sako) Arts Daily Supervisor: dr. V. (Vlado) Menkovski Graduation Committee: dr. V. (Vlado) Menkovski dr. D.C. (Decebal) Mocanu dr. N. (Nikolay) Yakovets May 16, 2018 v1.0 Abstract The focus of this thesis is dimensionality reduction of gene expression data. I propose and test a framework that deploys linear prediction algorithms resulting in a reduced set of selected genes relevant to a specified case. Abstract In cancer research there is a large need to automate parts of the process of diagnosis, this is mainly to reduce cost, make it faster and more accurate. -

Dopamine Response Gene Pathways in Dorsal Striatum Msns from a Gene
bioRxiv preprint doi: https://doi.org/10.1101/757500; this version posted September 5, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Dopamine response gene pathways in dorsal striatum MSNs from a gene 2 expression viewpoint: cAMP-mediated gene networks 3 Vladimir Babenko, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 630090, Novosibirsk 4 State University, Novosibirsk, Russia 630090; Anna Galyamina, Institute of Cytology and Genetics SB RAS, 5 Novosibirsk, Russia 630090; Igor Rogozin, National Institutes of Health, Rockville Pike, Bethesda, Maryland, 6 USA 20892; Dmitry Smagin, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 630090; 7 Natalia Kudryavtseva, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia 630090 8 9 *Corresponding author e-mail address: [email protected] (Vladimir Babenko) 10 Abstract 11 A mouse model of chronic social conflicts was used to analyze dorsal striatum neurons implicated in 12 cAMP-mediated phosphorylation activation pathways specific for Medium Spiny Neurons (MSNs). Based on 13 expression correlation analysis, we succeeded in dissecting Drd1- and Drd2-dopaminoceptive neurons (D1 14 and D2, correspondingly) gene pathways. We also found that D1 neurons feature previously reported two 15 states, passive and active ones, represented in our analysis by distinct, negatively correlated gene clusters. 16 The correlation based gene pathways strongly corroborate the phosphorylation cascades highlighted 17 in the previous studies, implying that the expression-based viewpoint corresponds to phosphorylation/ 18 dephosphorylation interplay in each type of neurons. Notably, D2 neurons showed the largest Ppp1r1b 19 (encoding DARPP-32) expression modulation impact, implying that Ppp1r1b expression dynamics is mostly 20 associated with neuroendocrine response mediated by Penk/Pdyn genes expression in D2 neurons. -

Genome-Wide Identification of Brain Mirnas in Response to High
Zhao et al. BMC Molecular Biol (2019) 20:3 https://doi.org/10.1186/s12867-019-0120-4 BMC Molecular Biology RESEARCH ARTICLE Open Access Genome‑wide identifcation of brain miRNAs in response to high‑intensity intermittent swimming training in Rattus norvegicus by deep sequencing Yanhong Zhao1*†, Anmin Zhang2,3*†, Yanfang Wang4, Shuping Hu3, Ruiping Zhang2 and Shuaiwei Qian2 Abstract Background: Physical exercise can improve brain function by altering brain gene expression. The expression mecha- nisms underlying the brain’s response to exercise still remain unknown. miRNAs as vital regulators of gene expres- sion may be involved in regulation of brain genes in response to exercise. However, as yet, very little is known about exercise-responsive miRNAs in brain. Results: We constructed two comparative small RNA libraries of rat brain from a high-intensity intermittent swim- ming training (HIST) group and a normal control (NC) group. Using deep sequencing and bioinformatics analysis, we identifed 2109 (1700 from HIST, 1691 from NC) known and 55 (50 from HIST, 28 from NC) novel candidate miRNAs. Among them, 34 miRNAs were identifed as signifcantly diferentially expressed in response to HIST, 16 were up- regulated and 18 were down-regulated. The results showed that all members of mir-200 family were strongly up- regulated, implying mir-200 family may play very important roles in HIST response mechanisms of rat brain. A total of 955 potential target genes of these 34 exercise-responsive miRNAs were identifed from rat genes. Most of them are directly involved in the development and regulatory function of brain or nerve. -

GWAS for Meat and Carcass Traits
G3: Genes|Genomes|Genetics Early Online, published on July 11, 2019 as doi:10.1534/g3.119.400452 GWAS for meat and carcass traits using imputed sequence level genotypes in pooled F2- designs in pigs Clemens Falker-Gieske,*,1,2 Iulia Blaj,†,2 Siegfried Preuß,‡ Jörn Bennewitz,‡ Georg Thaller,† Jens Tetens*,§ *Department of Animal Sciences, Georg-August-University, 37077 Göttingen, Germany. †Institute of Animal Breeding and Husbandry, Kiel University, 24118 Kiel, Germany. ‡Institute of Animal Husbandry and Breeding, University of Hohenheim, 70599 Stuttgart, Germany. §Center for Integrated Breeding Research, Georg-August-University, 37077 Göttingen, Germany. 1 © The Author(s) 2013. Published by the Genetics Society of America. Running title: Sequence level GWAS in pooled F2 pigs Keywords Genome wide association study Whole genome sequencing Imputation Meat, carcass, and production traits Variant calling 1Corresponding author: Clemens Falker-Gieske, Georg-August-University Goettingen, Department of Animal Sciences, Division Functional Breeding, Burckhardtweg 2, 37077 Goettingen, (+49) 551-39-23669 2contributed equally. 2 ABSTRACT In order to gain insight into the genetic architecture of economically important traits in pigs and to derive suitable genetic markers to improve these traits in breeding programs, many studies have been conducted to map quantitative trait loci. Shortcomings of these studies were low mapping resolution, large confidence intervals for quantitative trait loci-positions and large linkage disequilibrium blocks. Here, we overcome these shortcomings by pooling four large F2 designs to produce smaller linkage disequilibrium blocks and by resequencing the founder generation at high coverage and the F1 generation at low coverage for subsequent imputation of the F2 generation to whole genome sequencing marker density. -

SUPPLEMENTARY METHODS DNA Sequencing and Variant
Supplementary material J Med Genet SUPPLEMENTARY METHODS DNA sequencing and variant identification Genomic DNA was isolated from peripheral blood lymphocytes following standard procedures. Subsequently, exome enrichment was performed using the Agilent SureSelect Human All Exome V5 kit according to the manufacturer’s protocols. Exome sequencing was performed on an Illumina HiSeq system by BGI Europe (Copenhagen, Denmark). Read mapping along the hg19 reference genome (GRCh37/hg19) and variant calling were performed using BWA V.0.78(1) and GATK HaplotypeCaller V.3.3(2). A coverage of >20 reads was reached for 85.1% to 97.8% of the enriched regions. For variant annotation an in-house developed annotation and variant evaluation pipeline was used. For sequencing data of family W97-056, CNV detection was performed using CoNIFER V.0.2.2.(3) Genome sequencing was performed by BGI (Hong Kong, China) on a BGISeq500 using a 2x 100 bp paired end module, with a minimal median coverage of 30-fold per genome. Structural variants were called using Manta V.1.1.0(4) and CNVs using Control-FREEC.(5) Variants were validated and visualized using the IGV Software (V.2.4).(6) In the index case of family W08-1421, targeted DNA sequencing was performed using MIP sequencing.(7) MIPs were designed covering exons and exon-intron boundaries of a panel of 89 HL genes (Supplementary Table 6). Sequencing and data analysis were performed as previously described.(8) For each targeted region, an average coverage of 420 reads was obtained. A coverage of >20 reads was reached for 85.4% of the MIPs. -

Meta-Analysis of Gene Signatures and Key Pathways Indicates
www.nature.com/scientificreports OPEN Meta‑analysis of gene signatures and key pathways indicates suppression of JNK pathway as a regulator of chemo‑resistance in AML Parastoo Modarres1, Farzaneh Mohamadi Farsani1,3, Amir Abas Nekouie2 & Sadeq Vallian1* The pathways and robust deregulated gene signatures involved in AML chemo‑resistance are not fully understood. Multiple subgroups of AMLs which are under treatment of various regimens seem to have similar regulatory gene(s) or pathway(s) related to their chemo‑resistance phenotype. In this study using gene set enrichment approach, deregulated genes and pathways associated with relapse after chemotherapy were investigated in AML samples. Five AML libraries compiled from GEO and ArrayExpress repositories were used to identify signifcantly diferentially expressed genes between chemo‑resistance and chemo‑sensitive groups. Functional and pathway enrichment analysis of diferentially expressed genes was performed to assess molecular mechanisms related to AML chemotherapeutic resistance. A total of 34 genes selected to be diferentially expressed in the chemo‑ resistance compared to the chemo‑sensitive group. Among the genes selected, c-Jun, AKT3, ARAP3, GABBR1, PELI2 and SORT1 are involved in neurotrophin, estrogen, cAMP and Toll‑like receptor signaling pathways. All these pathways are located upstream and regulate JNK signaling pathway which functions as a key regulator of cellular apoptosis. Our expression data are in favor of suppression of JNK pathway, which could induce pro‑apoptotic gene expression as well as down regulation of survival factors, introducing this pathway as a key regulator of drug‑resistance development in AML. Acute myeloid leukemia (AML) is one of the most aggressive, life-threatening hematological malignancies char- acterized by uncontrolled proliferation of abnormal diferentiated and nonfunctional myeloid precursor cells1. -

UNIVERSITY of CALIFORNIA, IRVINE Epigenetic Control of Medial Habenula Function in Cocaine-Associated Behaviors DISSERTATION
UNIVERSITY OF CALIFORNIA, IRVINE Epigenetic control of medial habenula function in cocaine-associated behaviors DISSERTATION submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biological Sciences by Alberto J. López Dissertation Committee: Professor Marcelo A. Wood, Chair Assistant Professor Christie D. Fowler Assistant Professor Stephen V. Mahler 2018 © 2018 Alberto J. López Portion of Chapters 1 & 3 © 2018 John Wiley & Sons, Inc. Portion of Chapter 2 © 2016 The Journal of Neuroscience Dedicatoria A mi Abuelita Ethel quien me mostró lo que es ser fuerte, valiente, e inteligente To my wife Christina López of whom I am completely undeserving. Your love and support are the only reasons I’ve made it this far and I’m so incredibly lucky to have a true equal with whom to share this adventure. One sky, one destiny. To my family, Lorna, Jaime, Jaime Salvador, Cecilia, Elena, and the Burgarts who have all helped provide me with opportunities they never had. To the friends I’ve made along the way (including Drs. Andre White, Janine Kwapis, and Thekla Hemstedt, & Matt Saucedo, Rianne Campbell, Amni Al-Kachak, Matt Kwapis, Sarah Cross, and the Maddi) who have always provided incredible, and often thankless, support. To my various sources of caffeine throughout the Southland that have stimulated my life and work. “Simple, maybe, but not easy. There’s nothing easy about two men sharing one life...You see, sacrifice - that’s the price of a good trick.” Alfred Borden, The Prestige 1 ii Contents List of Figures ................................................................................................................................ iv Acknowledgments........................................................................................................................... v Curriculum Vitae ..........................................................................................................................