Capture and Molecular Control of Cd1a- Autoreactive T Cells in Human Skin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse
Welcome to More Choice CD Marker Handbook For more information, please visit: Human bdbiosciences.com/eu/go/humancdmarkers Mouse bdbiosciences.com/eu/go/mousecdmarkers Human and Mouse CD Marker Handbook Human and Mouse CD Marker Key Markers - Human Key Markers - Mouse CD3 CD3 CD (cluster of differentiation) molecules are cell surface markers T Cell CD4 CD4 useful for the identification and characterization of leukocytes. The CD CD8 CD8 nomenclature was developed and is maintained through the HLDA (Human Leukocyte Differentiation Antigens) workshop started in 1982. CD45R/B220 CD19 CD19 The goal is to provide standardization of monoclonal antibodies to B Cell CD20 CD22 (B cell activation marker) human antigens across laboratories. To characterize or “workshop” the antibodies, multiple laboratories carry out blind analyses of antibodies. These results independently validate antibody specificity. CD11c CD11c Dendritic Cell CD123 CD123 While the CD nomenclature has been developed for use with human antigens, it is applied to corresponding mouse antigens as well as antigens from other species. However, the mouse and other species NK Cell CD56 CD335 (NKp46) antibodies are not tested by HLDA. Human CD markers were reviewed by the HLDA. New CD markers Stem Cell/ CD34 CD34 were established at the HLDA9 meeting held in Barcelona in 2010. For Precursor hematopoetic stem cell only hematopoetic stem cell only additional information and CD markers please visit www.hcdm.org. Macrophage/ CD14 CD11b/ Mac-1 Monocyte CD33 Ly-71 (F4/80) CD66b Granulocyte CD66b Gr-1/Ly6G Ly6C CD41 CD41 CD61 (Integrin b3) CD61 Platelet CD9 CD62 CD62P (activated platelets) CD235a CD235a Erythrocyte Ter-119 CD146 MECA-32 CD106 CD146 Endothelial Cell CD31 CD62E (activated endothelial cells) Epithelial Cell CD236 CD326 (EPCAM1) For Research Use Only. -
![Cd1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719](https://docslib.b-cdn.net/cover/1446/cd1a-o10-concentrated-and-prediluted-monoclonal-antibody-901-3158-061719-341446.webp)
Cd1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719
CD1a [O10] Concentrated and Prediluted Monoclonal Antibody 901-3158-061719 Catalog Number: ACI 3158 A, B API 3158 AA VLTM 3158 G20 Description: 0.1, 0.5 mL conc. 6.0 mL, RTU 20 mL, RTU Dilution: 1:100 Ready-to-use Ready-to-use Diluent: Van Gogh Yellow N/A N/A Intended Use: Protocol Recommendations (VALENT® Automated Slide For In Vitro Diagnostic Use Staining Platform) Cont’d: CD1a [O10] is a mouse monoclonal antibody that is intended for Protein Block (Optional): Incubate for 10-20 minutes at RT with Val laboratory use in the qualitative identification of CD1a protein by Background Block. immunohistochemistry (IHC) in formalin-fixed paraffin-embedded Primary Antibody: Incubate for 30 minutes. (FFPE) human tissues. The clinical interpretation of any staining or its Secondary: Incubate for 10 minutes with Val Mouse Secondary. absence should be complemented by morphological studies using proper Linker: Incubate for 10 minutes with Val Universal Linker. controls and should be evaluated within the context of the patient’s Polymer: Incubate for 10 minutes with Val Universal Polymer. clinical history and other diagnostic tests by a qualified pathologist. Chromogen: Incubate for 5 minutes with Val DAB. Summary and Explanation: Counterstain: Counterstain for 5 minutes with Val Hematoxylin. CD1a is a protein of 43 to 49 kDa and is expressed on dendritic cells and cortical thymocytes (1,2). CD1a [O10] staining has been shown to be Protocol Recommendations (intelliPATH FLX® and manual use): useful in the differentiation of Langerhans cells from interdigitating cells. Peroxide Block: Block for 5 minutes with Peroxidazed 1. It has also proved useful for phenotyping Langerhans cell histiocytosis Pretreatment: Perform heat retrieval using Diva or Reveal Decloaker. -

Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma
ARTICLE https://doi.org/10.1038/s41467-020-16164-1 OPEN Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma Nayoung Kim 1,2,3,13, Hong Kwan Kim4,13, Kyungjong Lee 5,13, Yourae Hong 1,6, Jong Ho Cho4, Jung Won Choi7, Jung-Il Lee7, Yeon-Lim Suh8,BoMiKu9, Hye Hyeon Eum 1,2,3, Soyean Choi 1, Yoon-La Choi6,10,11, Je-Gun Joung1, Woong-Yang Park 1,2,6, Hyun Ae Jung12, Jong-Mu Sun12, Se-Hoon Lee12, ✉ ✉ Jin Seok Ahn12, Keunchil Park12, Myung-Ju Ahn 12 & Hae-Ock Lee 1,2,3,6 1234567890():,; Advanced metastatic cancer poses utmost clinical challenges and may present molecular and cellular features distinct from an early-stage cancer. Herein, we present single-cell tran- scriptome profiling of metastatic lung adenocarcinoma, the most prevalent histological lung cancer type diagnosed at stage IV in over 40% of all cases. From 208,506 cells populating the normal tissues or early to metastatic stage cancer in 44 patients, we identify a cancer cell subtype deviating from the normal differentiation trajectory and dominating the metastatic stage. In all stages, the stromal and immune cell dynamics reveal ontological and functional changes that create a pro-tumoral and immunosuppressive microenvironment. Normal resident myeloid cell populations are gradually replaced with monocyte-derived macrophages and dendritic cells, along with T-cell exhaustion. This extensive single-cell analysis enhances our understanding of molecular and cellular dynamics in metastatic lung cancer and reveals potential diagnostic and therapeutic targets in cancer-microenvironment interactions. 1 Samsung Genome Institute, Samsung Medical Center, Seoul 06351, Korea. -
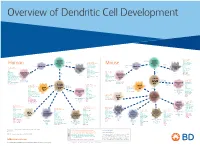
View Dendritic Cell Development Poster
Overview of Dendritic Cell Development Lineage–, CD45+, Common CD117 (c-kit) Common MHCII+, CD11c+ – + CD207 (Langerin) Myeloid CD117 (c-kit) Lineage , CD45 , Myeloid Progenitor MHCII (HLA-DR)+, CD11c+ Progenitor CD324 (E-Cadherin) Human Mouse CD326 (EpCAM) CD207 (Langerin) TGFb1 Cells CD11b, CD115 Cells CD14 Monocyte CD324 (E-Cadherin) Monocyte M-CSF CD11b – + Ly6C Langerhans CD24 Lineage , CD45 , M-CSF CD326 (EpCAM) MHCII (HLA-DR)+, CD11c+ Langerhans CD11blo Zbtb46– Cells CD172a (Sirp-α) CD16 CD1ahi, CD1c CD205 (DEC-205) Cells CSF F4/80 CD64 CD172a (Sirp-α) Lineage–, CD45+, FLT3L TLR3, TLR11 CD1a, CD1c Inflammatory CD369 (Dectin-1/CLEC7A) MHCII+, CD11c+ +/– CSF IL-15 CD8–, CD14– CD11b, CD14 CD371 (CLEC12A) CD64 Monocyte- FLT3L Inflammatory CD370 (Clec9a)– CD172a (Sirp-α) IL-15 CLEC6A CD11b derived lo Monocyte- CD206, CD209 (DC-SIGN) TLR1, TLR2, TLR3 , TLR6 CD209a (DC-SIGN) CD367 (DCIR/CLEC4A) DCs CD14– CD272 (BTLA)lo derived CD369 (Dectin-1/CLEC7A) DCs Common Ly-6C – + CD371 (CLEC12A) CD117 (c-kit) Lineage , CD45 , IL-1β, IL-6, IL-10, TLR1-6, TLR7-8, TLR10 Dendritic + lo CLEC6A – – CD135/FLT3 MHCII , CD11c IL-12, IL-23, TNF CD8a , CD14 IL-1β, IL-6 IL-10, Precursor TLR3lo, TLR4, TLR7, TLR8 CD45R (B220) IL-12, IL-23, TNF Plasmacytoid CD207 (Langerin)– Cells CD317 (BST-2) Common Lineage–, CD45+, FLT3L DCs Lineage–, CD45+, + Ly6C + lo/– CD207 IFN Type I + + Dendritic CD135/FLT3 MHCII (HLA-DR) , CD11c Lineage–, CD45+, IRF7, IRF8, BATF3hi Siglec-H MHCII (HLA-DR) , CD11c hi – + CD123 + + Dermal SpiB, Zbtb46 CD1a, CD64 CD1a Precursor CD117 (c-kit) -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -

Gene List HTG Edgeseq Immuno-Oncology Assay
Gene List HTG EdgeSeq Immuno-Oncology Assay Adhesion ADGRE5 CLEC4A CLEC7A IBSP ICAM4 ITGA5 ITGB1 L1CAM MBL2 SELE ALCAM CLEC4C DST ICAM1 ITGA1 ITGA6 ITGB2 LGALS1 MUC1 SVIL CDH1 CLEC5A EPCAM ICAM2 ITGA2 ITGAL ITGB3 LGALS3 NCAM1 THBS1 CDH5 CLEC6A FN1 ICAM3 ITGA4 ITGAM ITGB4 LGALS9 PVR THY1 Apoptosis APAF1 BCL2 BID CARD11 CASP10 CASP8 FADD NOD1 SSX1 TP53 TRAF3 BCL10 BCL2L1 BIRC5 CASP1 CASP3 DDX58 NLRP3 NOD2 TIMP1 TRAF2 TRAF6 B-Cell Function BLNK BTLA CD22 CD79A FAS FCER2 IKBKG PAX5 SLAMF1 SLAMF7 SPN BTK CD19 CD24 EBF4 FASLG IKBKB MS4A1 RAG1 SLAMF6 SPI1 Cell Cycle ABL1 ATF1 ATM BATF CCND1 CDK1 CDKN1B NCL RELA SSX1 TBX21 TP53 ABL2 ATF2 AXL BAX CCND3 CDKN1A EGR1 REL RELB TBK1 TIMP1 TTK Cell Signaling ADORA2A DUSP4 HES1 IGF2R LYN MAPK1 MUC1 NOTCH1 RIPK2 SMAD3 STAT5B AKT3 DUSP6 HES5 IKZF1 MAF MAPK11 MYC PIK3CD RNF4 SOCS1 STAT6 BCL6 ELK1 HEY1 IKZF2 MAP2K1 MAPK14 NFATC1 PIK3CG RORC SOCS3 SYK CEBPB EP300 HEY2 IKZF3 MAP2K2 MAPK3 NFATC3 POU2F2 RUNX1 SPINK5 TAL1 CIITA ETS1 HEYL JAK1 MAP2K4 MAPK8 NFATC4 PRKCD RUNX3 STAT1 TCF7 CREB1 FLT3 HMGB1 JAK2 MAP2K7 MAPKAPK2 NFKB1 PRKCE S100B STAT2 TYK2 CREB5 FOS HRAS JAK3 MAP3K1 MEF2C NFKB2 PTEN SEMA4D STAT3 CREBBP GATA3 IGF1R KIT MAP3K5 MTDH NFKBIA PYCARD SMAD2 STAT4 Chemokine CCL1 CCL16 CCL20 CCL25 CCL4 CCR2 CCR7 CX3CL1 CXCL12 CXCL3 CXCR1 CXCR6 CCL11 CCL17 CCL21 CCL26 CCL5 CCR3 CCR9 CX3CR1 CXCL13 CXCL5 CXCR2 MST1R CCL13 CCL18 CCL22 CCL27 CCL7 CCR4 CCRL2 CXCL1 CXCL14 CXCL6 CXCR3 PPBP CCL14 CCL19 CCL23 CCL28 CCL8 CCR5 CKLF CXCL10 CXCL16 CXCL8 CXCR4 XCL2 CCL15 CCL2 CCL24 CCL3 CCR1 CCR6 CMKLR1 CXCL11 CXCL2 CXCL9 CXCR5 -

Assessing Various Control Samples for Microarray Gene Expression Profiling of Laryngeal Squamous Cell Carcinoma
biomolecules Communication Assessing Various Control Samples for Microarray Gene Expression Profiling of Laryngeal Squamous Cell Carcinoma Adam Ustaszewski 1 , Magdalena Kostrzewska-Poczekaj 1, Joanna Janiszewska 1 , Malgorzata Jarmuz-Szymczak 1,2, Malgorzata Wierzbicka 1,3 , Joanna Marszal 3 , Reidar Grénman 4 and Maciej Giefing 1,* 1 Institute of Human Genetics, Polish Academy of Sciences, Strzeszy´nska32, 60-479 Pozna´n,Poland; [email protected] (A.U.); [email protected] (M.K.-P.); [email protected] (J.J.); [email protected] (M.J.-S.); [email protected] (M.W.) 2 Department of Oncology, Hematology and Bone Marrow Transplantation, Poznan University of Medical Sciences, 61-001 Pozna´n,Poland 3 Department of Otolaryngology and Laryngological Oncology, Pozna´nUniversity of Medical Sciences, 60-355 Pozna´n,Poland; [email protected] 4 Department of Otorhinolaryngology-Head and Neck Surgery, University of Turku and Turku University Hospital, 20520 Turku, Finland; seija.grenman@tyks.fi * Correspondence: maciej.giefi[email protected]; Tel.: +48-61-6579-138 Abstract: Selection of optimal control samples is crucial in expression profiling tumor samples. To address this issue, we performed microarray expression profiling of control samples routinely used in head and neck squamous cell carcinoma studies: human bronchial and tracheal epithelial cells, Citation: Ustaszewski, A.; squamous cells obtained by laser uvulopalatoplasty and tumor surgical margins. We compared Kostrzewska-Poczekaj, M.; the results using multidimensional scaling and hierarchical clustering versus tumor samples and Janiszewska, J.; Jarmuz-Szymczak, M.; laryngeal squamous cell carcinoma cell lines. A general observation from our study is that the Wierzbicka, M.; Marszal, J.; Grénman, R.; Giefing, M. -

Distribution Patterns of Dendritic Cells and T Cells in Diffuse Large B-Cell
Imaging, Diagnosis, Prognosis Distribution Patterns of Dendritic Cells and T Cells in Diffuse Large B-Cell Lymphomas Correlate with Prognoses Kung-Chao Chang,1, 2 Guan-Cheng Huang,3 DanJones,5 and Ya-Hui Lin4 Abstract Purpose: Diffuse large B-cell lymphoma (DLBCL), the most common subtype of non-Hodgkin’s lymphomas, accounts for 30% to 40% of all lymphoma cases. However, long-term survival by current chemotherapy was achieved in only 40% of patients, warranting the development of novel therapeutic strategies including T-cell immunotherapy. However, the level of baseline immune activation in DLBCL is unclear. Experimental Design: The density and distribution of dendritic cells and Tcells in 48 cases of primary DLBCL was evaluated by immunohistochemistry. Results: Increased numbers of intratumoral CD1a+ dendritic cells and increased S100+ cells and CD45RO+ T cells around the edges of the tumors were seen in 10 of 48 (21%), 9 of 48 (19%), and 10 of 48 (21%) cases and these were correlated with a favorable prognosis (P =0.015; P =0.070,andP = 0.017, respectively), along with increased granzyme B+ T cells in tumor beds (P = 0.013). Increased peritumoral T cells were correlated with tumor expression of HL A-DR (r = 0.446; P = 0.002). Extranodal lymphomas showed fewer tumor-associated CD45RO+ Tcells (r =-0.407;P = 0.001) and less conspicuous dendritic cell infiltrates. Conclusions: In DLBCL, the presence of baseline antitumor immune response is associated with favorable clinical outcome, and thus adjuvant T-cell immunotherapy may further boost treatment responses. The most common subtype of non-Hodgkin’s lymphoma is In animal models, the mice, which were immunized with diffuse large B-cell lymphoma (DLBCL; ref. -

Original Article the CD200/CD200R Mechanism in Mesenchymal Stem Cells’ Regulation of Dendritic Cells
Am J Transl Res 2021;13(8):9607-9613 www.ajtr.org /ISSN:1943-8141/AJTR0130885 Original Article The CD200/CD200R mechanism in mesenchymal stem cells’ regulation of dendritic cells Yulei Zhao, Guohong Su, Qing Wang, Ruihuan Wang, Minjuan Zhang The Second Department of Hematology, Cangzhou Central Hospital, 16 West Xinhua Road, Yunhe, Cangzhou, China Received January 27, 2021; Accepted April 12, 2021; Epub August 15, 2021; Published August 30, 2021 Abstract: Objective: To investigate the CD200/CD200R pathway mechanism in mesenchymal stem cells’ (MSC) regulation of dendritic cells (DC) (MSc). Methods: We collected marrow samples from 40 patients admitted to our hospital from January 2018 to December 2019. The bone marrow MSCs were cultivated, and the peripheral blood mononuclear cells (PBMC) and peripheral blood DC were isolated to establish an in vitro immune response model. The expressions of the CD200 molecule on the surface of MSC were measured. Anti-CD200 blocking antibodies were added to the culture system to observe the effect of the PBMC differentiation and the immature DC (imDC) to mature DC (mDC). Then the impact of the different positive rates of CD200 in the same MSC on imDC maturity was measured. Results: After adding mitogen pHA, the IL-4, IL-10, and TNF-α secretions were increased (all P<0.05), and the OD value of the PBMC+pHA group was higher than it was in the PBMC group. After stimulated by pHA, the CD200 of the MSC group was higher than it was in the MSC+PBMC group (P<0.05). The MSC+PBMC group co- culture inhibited the development of imDC to mDC. -

Table S3. RAE Analysis of Well-Differentiated Liposarcoma
Table S3. RAE analysis of well-differentiated liposarcoma Model Chromosome Region start Region end Size q value freqX0* # genes Genes Amp 1 145009467 145122002 112536 0.097 21.8 2 PRKAB2,PDIA3P Amp 1 145224467 146188434 963968 0.029 23.6 10 CHD1L,BCL9,ACP6,GJA5,GJA8,GPR89B,GPR89C,PDZK1P1,RP11-94I2.2,NBPF11 Amp 1 147475854 148412469 936616 0.034 23.6 20 PPIAL4A,FCGR1A,HIST2H2BF,HIST2H3D,HIST2H2AA4,HIST2H2AA3,HIST2H3A,HIST2H3C,HIST2H4B,HIST2H4A,HIST2H2BE, HIST2H2AC,HIST2H2AB,BOLA1,SV2A,SF3B4,MTMR11,OTUD7B,VPS45,PLEKHO1 Amp 1 148582896 153398462 4815567 1.5E-05 49.1 152 PRPF3,RPRD2,TARS2,ECM1,ADAMTSL4,MCL1,ENSA,GOLPH3L,HORMAD1,CTSS,CTSK,ARNT,SETDB1,LASS2,ANXA9, FAM63A,PRUNE,BNIPL,C1orf56,CDC42SE1,MLLT11,GABPB2,SEMA6C,TNFAIP8L2,LYSMD1,SCNM1,TMOD4,VPS72, PIP5K1A,PSMD4,ZNF687,PI4KB,RFX5,SELENBP1,PSMB4,POGZ,CGN,TUFT1,SNX27,TNRC4,MRPL9,OAZ3,TDRKH,LINGO4, RORC,THEM5,THEM4,S100A10,S100A11,TCHHL1,TCHH,RPTN,HRNR,FLG,FLG2,CRNN,LCE5A,CRCT1,LCE3E,LCE3D,LCE3C,LCE3B, LCE3A,LCE2D,LCE2C,LCE2B,LCE2A,LCE4A,KPRP,LCE1F,LCE1E,LCE1D,LCE1C,LCE1B,LCE1A,SMCP,IVL,SPRR4,SPRR1A,SPRR3, SPRR1B,SPRR2D,SPRR2A,SPRR2B,SPRR2E,SPRR2F,SPRR2C,SPRR2G,LELP1,LOR,PGLYRP3,PGLYRP4,S100A9,S100A12,S100A8, S100A7A,S100A7L2,S100A7,S100A6,S100A5,S100A4,S100A3,S100A2,S100A16,S100A14,S100A13,S100A1,C1orf77,SNAPIN,ILF2, NPR1,INTS3,SLC27A3,GATAD2B,DENND4B,CRTC2,SLC39A1,CREB3L4,JTB,RAB13,RPS27,NUP210L,TPM3,C1orf189,C1orf43,UBAP2L,HAX1, AQP10,ATP8B2,IL6R,SHE,TDRD10,UBE2Q1,CHRNB2,ADAR,KCNN3,PMVK,PBXIP1,PYGO2,SHC1,CKS1B,FLAD1,LENEP,ZBTB7B,DCST2, DCST1,ADAM15,EFNA4,EFNA3,EFNA1,RAG1AP1,DPM3 Amp 1 -

IT-Box-Systems for Flow-Cytometry
HOME IT-Box-Systems for Flow-Cytometry ImmunoTools special IT-Box-Systems give you the best price/quality ratio. The IT-Box-Systems designed for flow cytometry contain vials with each 100 µl of fluorescence conjugated antibodies against human antigens. anti-human human IT-Box 140: 100 fluorescence conjugated antibodies (FITC, PE, PerCP, PE/Dy647) anti-human human IT-Box 141: 100 fluorescence conjugated antibodies (FITC, PE, APC, PE/Dy647) and anti-mouse mouse IT-Box m278: 75 fluorescence conjugated antibodies (FITC, PE, APC) please note: The amount of antibodies you need for your assay is USER-dependent (dependent from cell amount, receptor density, incubation volume, incubation time, temperature, quality of the antibodies, quality of the laser and detectors .....). You can do between 10 to 25 experiments with each antibody. If you use the antibodies at the first time, please do some dilutions to find out the best concentration for your individual requirements. human IT-Box-140 100 fluorescence conjugated antibodies (small size, each 100 µl) against human surface antigens including Annexin V: Catalog# Price IT-Box-140 899 EUR contains FITC - conjugated anti- CD1a, CD2, CD3, CD4, CD8, CD10, CD11b, CD15, CD16, CD17, CD18, CD19, CD20, CD24, CD25, CD29, CD36, CD41a, CD45, CD45RO, CD46, CD52, CD53, CD54, CD56, CD58, CD61, CD62L, CD63, CD72, CD86, CD235ab, HLA-ABC, HLA-class II, Control-IgG1, Control-IgG2a, Control-IgG2b, Annexin V PE - conjugated anti- CD3, CD4, CD5, CD6, CD7, CD8, CD9, CD11a, CD11c, CD14, CD15, CD19, CD20, CD21, CD22, CD25, -

Human C3 Deficiency Associated with Impairments in Dendritic Cell
Human C3 Deficiency Associated with Impairments in Dendritic Cell Differentiation, Memory B Cells, and Regulatory T Cells This information is current as of September 27, 2021. Arije Ghannam, Martine Pernollet, Jean-Luc Fauquert, Nicole Monnier, Denise Ponard, Marie-Bernadette Villiers, Josette Péguet-Navarro, Arlette Tridon, Joel Lunardi, Denis Gerlier and Christian Drouet J Immunol 2008; 181:5158-5166; ; Downloaded from doi: 10.4049/jimmunol.181.7.5158 http://www.jimmunol.org/content/181/7/5158 http://www.jimmunol.org/ References This article cites 47 articles, 17 of which you can access for free at: http://www.jimmunol.org/content/181/7/5158.full#ref-list-1 Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 27, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2008 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal