(12) Patent Application Publication (10) Pub. No.: US 2012/0252869 A1 Collard Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Whole Exome Sequencing in Families at High Risk for Hodgkin Lymphoma: Identification of a Predisposing Mutation in the KDR Gene
Hodgkin Lymphoma SUPPLEMENTARY APPENDIX Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene Melissa Rotunno, 1 Mary L. McMaster, 1 Joseph Boland, 2 Sara Bass, 2 Xijun Zhang, 2 Laurie Burdett, 2 Belynda Hicks, 2 Sarangan Ravichandran, 3 Brian T. Luke, 3 Meredith Yeager, 2 Laura Fontaine, 4 Paula L. Hyland, 1 Alisa M. Goldstein, 1 NCI DCEG Cancer Sequencing Working Group, NCI DCEG Cancer Genomics Research Laboratory, Stephen J. Chanock, 5 Neil E. Caporaso, 1 Margaret A. Tucker, 6 and Lynn R. Goldin 1 1Genetic Epidemiology Branch, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 2Cancer Genomics Research Laboratory, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; 3Ad - vanced Biomedical Computing Center, Leidos Biomedical Research Inc.; Frederick National Laboratory for Cancer Research, Frederick, MD; 4Westat, Inc., Rockville MD; 5Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD; and 6Human Genetics Program, Division of Cancer Epidemiology and Genetics, National Cancer Institute, NIH, Bethesda, MD, USA ©2016 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2015.135475 Received: August 19, 2015. Accepted: January 7, 2016. Pre-published: June 13, 2016. Correspondence: [email protected] Supplemental Author Information: NCI DCEG Cancer Sequencing Working Group: Mark H. Greene, Allan Hildesheim, Nan Hu, Maria Theresa Landi, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Dilys Parry, Anand Pathak, Douglas R. Stewart, Philip R. Taylor, Geoffrey S. Tobias, Xiaohong R. Yang, Guoqin Yu NCI DCEG Cancer Genomics Research Laboratory: Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Amy A. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. -

Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared Amongst Δγ T, Innate Lymphoid, and Th Cells
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 δγ is online at: average * The Journal of Immunology , 10 of which you can access for free at: 2016; 197:1460-1470; Prepublished online 6 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600643 http://www.jimmunol.org/content/197/4/1460 Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst T, Innate Lymphoid, and Th Cells You Jeong Lee, Gabriel J. Starrett, Seungeun Thera Lee, Rendong Yang, Christine M. Henzler, Stephen C. Jameson and Kristin A. Hogquist J Immunol cites 41 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2016/07/06/jimmunol.160064 3.DCSupplemental This article http://www.jimmunol.org/content/197/4/1460.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 26, 2021. The Journal of Immunology Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst gd T, Innate Lymphoid, and Th Cells You Jeong Lee,* Gabriel J. -

Comparative Analysis of Chicken Chromosome 28 Provides New Clues to the Evolutionary Fragility of Gene-Rich Vertebrate Regions
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press Letter Comparative analysis of chicken chromosome 28 provides new clues to the evolutionary fragility of gene-rich vertebrate regions Laurie Gordon,1,2,5 Shan Yang,1,5 Mary Tran-Gyamfi,1,2 Dan Baggott,1 Mari Christensen,1,2 Aaron Hamilton,1 Richard Crooijmans,3 Martien Groenen,3 Susan Lucas,2 Ivan Ovcharenko,2,4 and Lisa Stubbs1,6 1Genome Biology Group, Lawrence Livermore National Laboratory, Livermore, California 94550, USA; 2Department of Energy Joint Genome Institute, Walnut Creek, California 94598, USA; 3Wageningen University, Wageningen 6709 PG, The Netherlands; 4Computations Group, Lawrence Livermore National Laboratory, Livermore, California 94550, USA The chicken genome draft sequence has provided a valuable resource for studies of an important agricultural and experimental model species and an important data set for comparative analysis. However, some of the most gene-rich segments are missing from chicken genome draft assemblies, limiting the analysis of a substantial number of genes and preventing a closer look at regions that are especially prone to syntenic rearrangements. To facilitate the functional and evolutionary analysis of one especially gene-rich, rearrangement-prone genomic region, we analyzed sequence from BAC clones spanning chicken microchromosome GGA28; as a complement we also analyzed a gene-sparse, stable region from GGA11. In these two regions we documented the conservation and lineage-specific gain and loss of protein-coding genes and precisely mapped the locations of 31 major human-chicken syntenic breakpoints. Altogether, we identified 72 lineage-specific genes, many of which are found at or near syntenic breaks, implicating evolutionary breakpoint regions as major sites of genetic innovation and change. -

0Nducing *Nd Suppressing Phe *5Pern*Pive 5Engphening Of
nducing nd suppressing he ern)ive enghening of eomeres mech)nism in cncer ce0s Dissertation submitted by Delia Braun 2018 Dissertation submitted to the Combined Faculties for the Natural Sciences and for Mathematics of the Ruperto-Carola University of Heidelberg, Germany for the degree of Doctor of Natural Sciences presented by Dipl.-Chem. Delia Maria Braun born in Offenburg, Germany Oral examination: 24.07.2018 nducing nd suppressing he ern)ive enghening of eomeres mech)nism in cncer ce0s Referees: Prof. Dr. Karsten Rippe Prof. Dr. Brian Luke This work was performed from September 2012 to January 2018 under the supervision of Prof. Dr. Karsten Rippe in the Division Chromatin Networks at the DKFZ and the Bioquant Center in Heidelberg, Germany. Declaration I hereby declare that I have written the submitted dissertation “Inducing and suppressing the alternative lengthening of telomeres mechanism in cancer cells” myself and in this process, have used no other sources or materials than those explicitly indicated. I hereby declare that I have not applied to be examined at any other institution, nor have I used the dissertation in this or any other form at any other institution as an examination paper, nor submitted it to any other faculty as a dissertation. ______________________________ ______________________________ (Place, Date) Delia Braun Für Philipp und Lina Tbe of conens Lis of pubicions .................................................. V Summ)r? ............................................................... V! Zusmmenfssung ................................................ -

An Integrative Genomic Analysis of the Longshanks Selection Experiment for Longer Limbs in Mice
bioRxiv preprint doi: https://doi.org/10.1101/378711; this version posted August 19, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Title: 2 An integrative genomic analysis of the Longshanks selection experiment for longer limbs in mice 3 Short Title: 4 Genomic response to selection for longer limbs 5 One-sentence summary: 6 Genome sequencing of mice selected for longer limbs reveals that rapid selection response is 7 due to both discrete loci and polygenic adaptation 8 Authors: 9 João P. L. Castro 1,*, Michelle N. Yancoskie 1,*, Marta Marchini 2, Stefanie Belohlavy 3, Marek 10 Kučka 1, William H. Beluch 1, Ronald Naumann 4, Isabella Skuplik 2, John Cobb 2, Nick H. 11 Barton 3, Campbell Rolian2,†, Yingguang Frank Chan 1,† 12 Affiliations: 13 1. Friedrich Miescher Laboratory of the Max Planck Society, Tübingen, Germany 14 2. University of Calgary, Calgary AB, Canada 15 3. IST Austria, Klosterneuburg, Austria 16 4. Max Planck Institute for Cell Biology and Genetics, Dresden, Germany 17 Corresponding author: 18 Campbell Rolian 19 Yingguang Frank Chan 20 * indicates equal contribution 21 † indicates equal contribution 22 Abstract: 23 Evolutionary studies are often limited by missing data that are critical to understanding the 24 history of selection. Selection experiments, which reproduce rapid evolution under controlled 25 conditions, are excellent tools to study how genomes evolve under strong selection. Here we 1 bioRxiv preprint doi: https://doi.org/10.1101/378711; this version posted August 19, 2018. -

PDZD7-MYO7A Complex Identified in Enriched Stereocilia Membranes
RESEARCH ARTICLE PDZD7-MYO7A complex identified in enriched stereocilia membranes Clive P Morgan1†, Jocelyn F Krey1†, M’hamed Grati2, Bo Zhao3, Shannon Fallen1, Abhiraami Kannan-Sundhari2, Xue Zhong Liu2, Dongseok Choi4,5, Ulrich Mu¨ ller3, Peter G Barr-Gillespie1* 1Oregon Hearing Research Center and Vollum Institute, Oregon Health and Science University, Portland, United States; 2Department of Otolaryngology, Miller School of Medicine, University of Miami, Miami, United States; 3Dorris Neuroscience Center, The Scripps Research Institute, La Jolla, United States; 4OHSU-PSU School of Public Health, Oregon Health and Science University, Portland, United States; 5Graduate School of Dentistry, Kyung Hee University, Seoul, Korea Abstract While more than 70 genes have been linked to deafness, most of which are expressed in mechanosensory hair cells of the inner ear, a challenge has been to link these genes into molecular pathways. One example is Myo7a (myosin VIIA), in which deafness mutations affect the development and function of the mechanically sensitive stereocilia of hair cells. We describe here a procedure for the isolation of low-abundance protein complexes from stereocilia membrane fractions. Using this procedure, combined with identification and quantitation of proteins with mass spectrometry, we demonstrate that MYO7A forms a complex with PDZD7, a paralog of USH1C and DFNB31. MYO7A and PDZD7 interact in tissue-culture cells, and co-localize to the ankle-link region of stereocilia in wild-type but not Myo7a mutant mice. Our data thus describe a new paradigm for *For correspondence: gillespp@ the interrogation of low-abundance protein complexes in hair cell stereocilia and establish an ohsu.edu unanticipated link between MYO7A and PDZD7. -
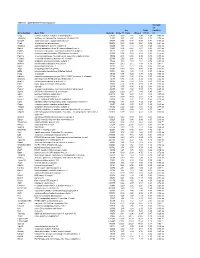
Table S-1 Mpkccd Transcriptome
Table S1. mpkCCD Cell Transcriptomes. Correlati on Ori. Ratio Coefficie Gene Symbol Gene Title GeneID Clone 11 Clone Clone 2 (11/2) nt Copg coatomer protein complex, subunit gamma 54161 1.99 1.81 2.48 0.80 -0.49 ns Atp6v0d1 ATPase, H+ transporting, lysosomal V0 subunit D1 11972 3.37 2.87 3.00 1.12 0.74 ns Golga7 golgi autoantigen, golgin subfamily a, 7 57437 3.03 3.40 3.58 0.84 -0.34 ns Psph phosphoserine phosphatase 100678 0.38 0.66 0.64 0.59 -0.61 ns Trappc4 trafficking protein particle complex 4 60409 1.17 1.12 1.08 1.08 0.64 ns Dpm2 dolichol-phosphate (beta-D) mannosyltransferase 2 13481 0.74 0.87 0.81 0.91 -0.31 ns Psmb5 proteasome (prosome, macropain) subunit, beta type 5 19173 2.92 3.00 2.92 0.99 0.32 ns Dhrs1 dehydrogenase/reductase (SDR family) member 1 52585 0.74 0.68 0.88 0.83 -0.18 ns Ppm1a protein phosphatase 1A, magnesium dependent, alpha isoform 19042 2.43 2.60 2.96 0.82 -0.53 ns Psenen presenilin enhancer 2 homolog (C. elegans) 66340 2.52 2.88 3.80 0.66 -0.77 ns Anapc1 anaphase promoting complex subunit 1 17222 1.16 1.33 1.61 0.72 -0.33 ns Mrpl43 mitochondrial ribosomal protein L43 94067 2.15 2.15 1.84 1.16 0.91 * Nmt1 N-myristoyltransferase 1 18107 3.21 2.71 3.36 0.95 0.31 ns Atg5 autophagy-related 5 (yeast) 11793 0.70 0.73 0.66 1.05 -0.23 ns Mtif2 mitochondrial translational initiation factor 2 76784 1.16 1.29 1.19 0.97 -0.36 ns Psap prosaposin 19156 8.07 7.25 8.37 0.96 0.02 ns Ube2g1 ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, C. -

4 353 Skin Oral 1 B
A B Supplementary Figure S1: Differentially expressed piRNAs during skin and oral mucosal wound healing. (A) piRNA 0hr-1 0hr-3 0hr-2 24hr-2 24hr-1 24hr-3 5day-3 5day-1 5day-2 0hr-1 0hr-2 0hr-3 24hr-2 24hr-1 24hr-3 5day-1 5day-2 5day-3 profiles were obtained on mouse skin and oral mucosal (palate) wound healing time course (0hr, 24 hr, and 5 day). A total of 357 differentially expressed piRNA were identified during skin wound healing (Bonferroni adjusted P value <0.05). See Supplementary Table 1A for the full list. C (B) Five differentially expressed piRNA skin were identified during oral mucosal wound healing (P value <0.01, list presented in Supplementary Table 1B). Note: more 353 stringent statistical cut-off ((Bonferroni adjusted P value) yield 0 differentially expressed piRNA gene. (C) Venn diagram illustrates overlaps between differentially 4 expressedpiRNAsinskinandoral oral 1 mucosal wound healing. min max Supplementary Table S1a: Differentially expressed piRNAs in skin wound healing Mean StDev piRNA 0 hr 24 hr 5 day 0 hr 24 hr 5 day pVal adj P piR‐mmu‐15927330 5.418351 11.39746 10.799 0.34576 0.253492 0.154802 6.32E‐14 6.97E‐11 piR‐mmu‐49559417 5.301647 10.10777 9.816878 0.441719 0.222335 0.032479 3.95E‐12 4.36E‐09 piR‐mmu‐30053093 6.32531 11.26384 4.020902 0.280841 1.057847 0.178798 5.21E‐12 5.76E‐09 piR‐mmu‐29303577 5.15005 10.47662 9.52175 0.554877 0.26622 0.163283 1.53E‐11 1.69E‐08 piR‐mmu‐49254706 5.187673 10.19644 9.622374 0.520671 0.330378 0.191116 1.96E‐11 2.16E‐08 piR‐mmu‐49005170 5.415133 9.639725 9.565967 0.411507 0.281143 -

You Can Check If Genes Are Captured by the Agilent Sureselect V5 Exome
IIHG Clinical Exome Sequencing Test Gene Coverage • Whole Exome Sequencing is a targeted capture platform which does not capture the entire exome. Regions not captured by the exome will not be analyzed. o Please note, it is important to understand the absence of a reportable variant in a given gene does not mean there are not pathogenic variants in that gene. • Data sensitivity and specificity for exome testing is variable as gene coverage is not uniform throughout the exome. • The Agilent SureSelect XT Human All Exon v5 kit captures ~98% of Refseq coding base pairs, and >94% of the captured coding bases in the exome are covered at our depth-of-coverage minimum threshold (30 reads). • All test reports include the following information: o Regions of the Symptom Candidate Gene List which were not captured by the current exome capture platform. o Regions of the Symptom Candidate Gene List which were not sequenced to a sufficient depth of coverage to make a clinical diagnosis. • You can check if genes are captured by the Agilent Agilent SureSelect v5 exome capture, and how well those genes are typically covered here. • Explanations for the headings; • Symbol: Gene symbol • Chr: Chromosome • % Captured: the minimum portion of the gene captured by the Agilent SureSelect XT Human All Exon v5 kit. This includes all transcripts for a given gene. o 100.00% = the entire gene is captured o 0.00% = none of the gene is captured • % Covered: the minimum portion of the gene that has at least 30x average coverage when sequenced on the Illumina HiSeq2000 with 100 base pair (bp) paired-end reads for eight CEPH samples.