Hybridization Hotspots at Bat Swarming Sites
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

2010-7-Alcathoe's
An awful lot of bat workers are going to be looking even Alcathoe Bat more closely at any Myotis they encounter. You had to be there Top photo (c) http://www.krzysztof.piksa-pl.com As we arrived at the ZSL symposium people were in little Lower photo Derek Smith’s mystery bat huddles muttering excitedly. Soon we were as excited Prof John Altringham was using the symposium to announce that Britain has a new bat species The news was kept quiet until the results had been checked and double checked but there can be no doubt. We have discussed before the nightmare which is distinguishing Brandt and whiskered bats. One of John’s research students was doing genetic analysis of Brandt and Whiskered bats and contacted John because she was getting a sizeable cluster of anomalous results that fitted neither species when she looked at the gene sequences. DNA was taken from bats in the North York Moors and Susses, and when they also looked at samples from Europe, they got the same results John says that key features to look for are forearm length, and echolocation calls. Bob and Jude were insufferably smug having seen it in Hungary ad knew how to pronounce its name (Al Kath- oh-ee). John is in favour of calling the bat Alcathoe Bat rather than Alcathoe’s John kindly sent us a copy of his draft paper. We forwarded this to taxonomy keeny Derek Smith and we could almost hear him weeping as he e mailed us back post-haste “Hi Bob and Jude, many thanks for the paper. -

Index of Handbook of the Mammals of the World. Vol. 9. Bats
Index of Handbook of the Mammals of the World. Vol. 9. Bats A agnella, Kerivoula 901 Anchieta’s Bat 814 aquilus, Glischropus 763 Aba Leaf-nosed Bat 247 aladdin, Pipistrellus pipistrellus 771 Anchieta’s Broad-faced Fruit Bat 94 aquilus, Platyrrhinus 567 Aba Roundleaf Bat 247 alascensis, Myotis lucifugus 927 Anchieta’s Pipistrelle 814 Arabian Barbastelle 861 abae, Hipposideros 247 alaschanicus, Hypsugo 810 anchietae, Plerotes 94 Arabian Horseshoe Bat 296 abae, Rhinolophus fumigatus 290 Alashanian Pipistrelle 810 ancricola, Myotis 957 Arabian Mouse-tailed Bat 164, 170, 176 abbotti, Myotis hasseltii 970 alba, Ectophylla 466, 480, 569 Andaman Horseshoe Bat 314 Arabian Pipistrelle 810 abditum, Megaderma spasma 191 albatus, Myopterus daubentonii 663 Andaman Intermediate Horseshoe Arabian Trident Bat 229 Abo Bat 725, 832 Alberico’s Broad-nosed Bat 565 Bat 321 Arabian Trident Leaf-nosed Bat 229 Abo Butterfly Bat 725, 832 albericoi, Platyrrhinus 565 andamanensis, Rhinolophus 321 arabica, Asellia 229 abramus, Pipistrellus 777 albescens, Myotis 940 Andean Fruit Bat 547 arabicus, Hypsugo 810 abrasus, Cynomops 604, 640 albicollis, Megaerops 64 Andersen’s Bare-backed Fruit Bat 109 arabicus, Rousettus aegyptiacus 87 Abruzzi’s Wrinkle-lipped Bat 645 albipinnis, Taphozous longimanus 353 Andersen’s Flying Fox 158 arabium, Rhinopoma cystops 176 Abyssinian Horseshoe Bat 290 albiventer, Nyctimene 36, 118 Andersen’s Fruit-eating Bat 578 Arafura Large-footed Bat 969 Acerodon albiventris, Noctilio 405, 411 Andersen’s Leaf-nosed Bat 254 Arata Yellow-shouldered Bat 543 Sulawesi 134 albofuscus, Scotoecus 762 Andersen’s Little Fruit-eating Bat 578 Arata-Thomas Yellow-shouldered Talaud 134 alboguttata, Glauconycteris 833 Andersen’s Naked-backed Fruit Bat 109 Bat 543 Acerodon 134 albus, Diclidurus 339, 367 Andersen’s Roundleaf Bat 254 aratathomasi, Sturnira 543 Acerodon mackloti (see A. -

Chiropterology Division BC Arizona Trial Event 1 1. DESCRIPTION: Participants Will Be Assessed on Their Knowledge of Bats, With
Chiropterology Division BC Arizona Trial Event 1. DESCRIPTION: Participants will be assessed on their knowledge of bats, with an emphasis on North American Bats, South American Microbats, and African MegaBats. A TEAM OF UP TO: 2 APPROXIMATE TIME: 50 minutes 2. EVENT PARAMETERS: a. Each team may bring one 2” or smaller three-ring binder, as measured by the interior diameter of the rings, containing information in any form and from any source. Sheet protectors, lamination, tabs and labels are permitted in the binder. b. If the event features a rotation through a series of stations where the participants interact with samples, specimens or displays; no material may be removed from the binder throughout the event. c. In addition to the binder, each team may bring one unmodified and unannotated copy of either the National Bat List or an Official State Bat list which does not have to be secured in the binder. 3. THE COMPETITION: a. The competition may be run as timed stations and/or as timed slides/PowerPoint presentation. b. Specimens/Pictures will be lettered or numbered at each station. The event may include preserved specimens, skeletal material, and slides or pictures of specimens. c. Each team will be given an answer sheet on which they will record answers to each question. d. No more than 50% of the competition will require giving common or scientific names. e. Participants should be able to do a basic identification to the level indicated on the Official List. States may have a modified or regional list. See your state website. -
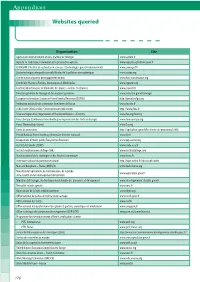
Appendices Websites Queried
Appendices Websites queried Organization Site Agence de l’environnement et de la maîtrise de l’énergie www.ademe.fr Agreste: la statistique, l’évaluation et la prospective agricole www.agreste.agriculture.gouv.fr CEMAGREF (l’Institut de recherche en sciences et technologies pour l’environnement) www.cemagref.fr Centre technique interprofessionnel d’études de la pollution atmosphérique www.citepa.org Comité national pour le développement du bois www.bois-construction.org Comité des Plantes à Parfum, Aromatiques et Médicinales www.cpparm.org Confédération française de l’industrie des papiers, cartons et celluloses www.copacel.fr Direction générale de l’énergie et des matières premières www.industrie.gouv.fr/energie European Information System on Forest Genetic Resources (EUFGIS) http://portal.eufgis.org Fédération nationale des communes forestières de France www.fncofor.fr FCBA (Forêt Cellulose Bois-Construction Ameublement) http://www.fcba.fr Food and Agriculture Organization of the United Nations – Forestry www.fao.org/forestry Forest Europe (Conférence ministérielle pour la protection des forêts en Europe) www.foresteurope.org Forest Stewardship Council www.fsc.org Forêts de protection http://agriculture.gouv.fr/les-forets-de-protection,10806 French National Forest Inventory (Inventaire forestier national) www.ifn.fr Groupement d’intérêt public Écosystèmes Forestiers www.gip-ecofor.org Institut de l’abeille (ITSAP) www.cnda.asso.fr Institut méditerranéen du liège (IML) www.institutduliege.com Institut national de la statistique et des -

Hungary and Slovakia, 2017
HUNGARY and SLOVAKIA SMALL MAMMAL TOUR - The Bats and Rodents of Central Europe Hangarian hay meadow in warm August sunshine. Steve Morgan ([email protected]), John Smart 25/8/17 HUNGARY and SLOVAKIA SMALL MAMMAL TOUR 1 Introduction I had long intended to visit Hungary for bats and small mammals but had never quite got round to it. Now, however, a chance presented itself to join a tour with both Hungary and Slovakia on the itinerary and a long list of prospective mammalian targets on offer, including Forest Dormouse, European Hamster, Lesser Mole Rat, Common Souslik and a number of highly desirable bats such as Grey Long-eared, Northern and Parti-coloured. The tour was organised by Ecotours of Hungary and led by Istvan Bartol. It ran from 9/8/17 to 17/8/17, the two particpants being John Smart and me, both of us from the UK. 2 Logistics I flew from Luton to Budapest on Wizzair. Frankly, I’d never heard of Wizzair before and, given their two hour delay on the outward leg (resulting in an extremely late check in to my hotel in Budapest), I’m not sure I want to hear about them again! The hotels selected by Ecotours were all very good. In Mezokovesd we stayed at the Hajnal Hotel which was clean and comfortable and offered a good (cooked) buffet breakfast. In Slovakia we stayed at the equally good Penzion Reva which was set in very nice countryside overlooking a picturesque lake. Istvan Bartol led the tour and did all the driving. -

Identification of Bats in Hibernation
Pierrette Nyssen and Ruddy Cors Plecotus / Natagora Octobre 2019 David Galens Vespertilio vzw May 2020 Identification of bats in hibernation 1 October 2019 Département : Plecotus, pôle chauves-souris de Natagora asbl Traverse des Muses 1 5000 Namur Belgique [email protected] - www.chauves-souris.be [email protected] - www.natagora.be www.vespertilio.be Editors: Pierrette Nyssen - [email protected] Lay-out: Ruddy Cors - [email protected] With the cooperation of: Jean-Louis Gathoye, Anne-Catherine Martin, Gilles San Martin, Quentin Smits, Nicolas Titeux and all the illustrators and photographers named in the document. Cover photo: Ruddy Cors (Myotis emarginatus) English translation: David Galens – Vespertilio vzw – May 2020 – www.vespertilio.be. A special thanks to John Haddow, Stuart Newson and Nils Bouillard for their linguistic input. All the photoghraphs mentioning Dietz & von Helversen are used with agreement of the author, Dietz, C. & von Helversen, O. 2004. Identification key to the bats of Europe, 72 pp., version 1.0 - electronical publication downloadable on the author’s website: http://www.fledermausdietz.de/publications/publications.html All the illustrations mentioning of Punt et al. are taken from the book Punt, A., Van Bree, P. J. H., De Vlas, J. & Wiersema, G.J., 1974. De Nederlandse vleermuizen, Wetenschappelijke mededelingen KNNV 104: 48 pg. It is the goal for this guide to evolve. If you have any comments about the proposed identification characteristics or know of other (reliable) methods, feel free to share those with us at [email protected]. This guide is constantly updated following comments and suggestions. You can always download the latest version via www.chauves-souris.be: under "Publications" - "Documents à télécharger" or www.vespertilio.be. -

On the Distribution and Taxonomy of Bats of the Myotis Mystacinus Morphogroup from the Caucasus Region (Chiroptera: Vespertilionidae)
Turkish Journal of Zoology Turk J Zool (2016) 40: 842-863 http://journals.tubitak.gov.tr/zoology/ © TÜBİTAK Research Article doi:10.3906/zoo-1505-47 On the distribution and taxonomy of bats of the Myotis mystacinus morphogroup from the Caucasus region (Chiroptera: Vespertilionidae) 1,2, 3 4,5 Petr BENDA *, Suren GAZARYAN , Peter VALLO 1 Department of Zoology, National Museum (Natural History), Prague, Czech Republic 2 Department of Zoology, Faculty of Science, Charles University in Prague, Prague, Czech Republic 3 UNEP/EUROBATS Secretariat, United Nations Campus, Bonn, Germany 4 Institute of Vertebrate Biology, Academy of Sciences of the Czech Republic, Brno, Czech Republic 5 Institute of Evolutionary Ecology and Conservation Genomics, Ulm University, Ulm, Germany Received: 29.05.2015 Accepted/Published Online: 18.12.2015 Final Version: 06.12.2016 Abstract: Due to the combination of general morphological similarity among representatives of particular populations and/or taxa, the taxonomy of the Myotis mystacinus morphogroup is traditionally considered to be one of the most complex topics of bat systematics. We used a combination of genetic and morphological approaches to identify which taxa of the morphogroup inhabit the broader Caucasus region (i.e. the territory between Crimea, Southern Russia, Northern Anatolia, and Northern Iran, incl.) and to define the relationships of these taxa to other Palaearctic populations of the morphogroup. Based on the genetic identification (complete sequence of the cytochrome b gene) of 56 specimens of the morphogroup originating from the region and the subsequent morphological comparison of more than 100 museum specimens, five species were shown to occur in the Caucasus region: Myotis mystacinus s.str., M. -

Manual of Sonochiro® 3.0
recherche & développement recherche & développement 1 Manual of SonoChiro® 3.0 I) Introduction I.1) Purpose and framework for the software SonoChiro® is a software analysing automatically ultrasound recordings of bats, be it in time expansion or in real time. This software provides a valuable help for all users of bat detectors, in particular in case of unattended recorders accumulating large amounts of data. Its diverse functions make it useful in many different contexts of analysis: in- ventorying species at a site, counting bat passes, quantifying hunting activity (feeding buzzes), highlighting social activity, etc. SonoChiro® cannot completely substitute for a manual identification since a zero er- ror rate is unattainable in the current state of knowledge and technological advances. However, SonoChiro® has been designed for maximum ease of the treatment of its own mistakes, especially through a set of relevant confidence indices. Thus, the time saved through the use of SonoChiro® is considerable. This varies depending on many factors (objectives, recording quality, etc.) but it is generally greater than 90%, it even exceeds 99% in many cases. Copyright © Biotope, Research & Development, 2013 • Biotope Society - BP58 - 34140 Mèze - France - www.biotope.fr 2 I.2) Principle of operation SonoChiro® process recordings in two phases: detection and classification. The detection process consists in localizing then characterizing a maximum number of signals po- tentially emitted by bats. SonoChiro® has indeed been designed to ensure a maximum sensitivity in bat calls detection, in order to detect even the most discrete species, even in case of weak signals and/or poor quality recordings. This approach result in large numbers of parasites being, in this preliminary phase, considered as potential bat calls, but these are easily discarded during the next phase (see also par IV, sensitivity settings). -

2.2 Binsted Woods Bat Trapping Survey by Daniel Whitby of Animal Ecology & Wildlife Consultants
AEWC Ltd Animal Ecology & Wildlife Consultants Butlers House North Street Petworth West Sussex GU28 0DP Tel: +44(0)8452 505 585 Mob: +44(0)7764 813002 e-mail: [email protected] Bat Survey Trapping Survey Binsted Woods MAVES Group Arundel Daniel Whitby 16-012-BS 2016 Registered Office: Butlers House, North Street, Petworth, West Sussex, GU28 9DP Registered in England and Wales No. 06527840 AEWC Ltd This report has been prepared by AEWC Limited, with all reasonable skill, care and diligence within the terms of the Contract with the client. We disclaim any responsibility to the client and others in respect of any matters outside the scope of the above. This report is confidential to the client and we accept no responsibility of whatsoever nature to third parties to whom this report, or any part thereof, is made known. Any such party relies on the report at their own risk. 1 AEWC Ltd 1 Introduction 1.1 This Bat survey and report has been carried out and prepared by Daniel Whitby, an English Nature Licensed bat worker. The survey involved using trapping surveys across the site with an acoustic lure playing a range of species social calls to trap bats present on the site to identify species, sex and breeding status. 1.2 The objective of this survey was to conduct an initial baseline survey to indicate the species assemblages, however particular interest was aimed at identifying any notably rare species and if necessary radio tagging to identify roost locations of any notably rare species to inform on roost location and hence the site importance. -

Brandt's Bat (Myotis Brandtii)
European Community Directive on the Conservation of Natural Habitats and of Wild Fauna and Flora (92/43/EEC) Fourth Report by the United Kingdom under Article 17 on the implementation of the Directive from January 2013 to December 2018 Supporting documentation for the conservation status assessment for the species: S1320 ‐ Brandt's bat (Myotis brandtii) ENGLAND IMPORTANT NOTE ‐ PLEASE READ • The information in this document is a country‐level contribution to the UK Reporton the conservation status of this species, submitted to the European Commission aspart of the 2019 UK Reporting under Article 17 of the EU Habitats Directive. • The 2019 Article 17 UK Approach document provides details on how this supporting information was used to produce the UK Report. • The UK Report on the conservation status of this species is provided in a separate doc‐ ument. • The reporting fields and options used are aligned to those set out in the European Com‐ mission guidance. • Explanatory notes (where provided) by the country are included at the end. These pro‐ vide an audit trail of relevant supporting information. • Some of the reporting fields have been left blank because either: (i) there was insuffi‐ cient information to complete the field; (ii) completion of the field was not obligatory; (iii) the field was not relevant to this species (section 12 Natura 2000 coverage forAnnex II species) and/or (iv) the field was only relevant at UK‐level (sections 9 Future prospects and 10 Conclusions). • For technical reasons, the country‐level future trends for Range, Population and Habitat for the species are only available in a separate spreadsheet that contains all the country‐ level supporting information. -

One of UK's Rarest Bats Spotted in Wiltshire Woods 22 August 2013
One of UK's rarest bats spotted in Wiltshire woods 22 August 2013 landscape to benefit this rare bat and other species." Classified as Vulnerable by the 2000 IUCN Red List, barbastelles are identified by their long white- tipped, dark-brown fur which gives them an almost frosty look. They typically live in wooded areas near water. These skilled hunters will emerge as the sun sets and hunt for moths and flies. A barbastelle in flight. Credit: Dietmar Nill Ashley White said: "The sighting of a barbastelle tells us that the woodland is well-managed so it is really good news for the Trust. Through the summer, these bats will roost in trees and the bat During a night of bat trapping on Wiltshire Wildlife boxes placed in the woods by the Trust, and will Trust's Blackmoor Copse nature reserve, Phil occasionally also roost in crevices in buildings. As Brown, an MSc student at the University of Bristol, summer fades into autumn, barbastelles will be identified a barbastelle bat. This is the first mating and, fingers crossed, next summer confirmed sighting of this elusive and rare species Blackmoor Copse will have a new generation of of bat on the reserve. barbastelles!" Working with the Trust's reserve officer, Ashley Phil Brown added: "The barbastelle was a great White, Phil set up traps to discover what species catch and record but we also caught two other bats call the woods of Blackmoor Copse home. of interest. My research is focussing on finding a species of bat called the Alcathoe bat that was only Phil Brown said: "It's great to catch a rare bat such described as a species new to science as recently as a barbastelle. -

Agreement for the Conservation of Bats in Europe (Eurobats)
Inf.EUROBATS.MoP7.43 AGREEMENT FOR THE CONSERVATION OF BATS IN EUROPE (EUROBATS) Report on the implementation of the Agreement in the United Kingdom 2014 This document reports on actions undertaken by the United Kingdom (UK) between January 2010 and December 2013 to meet its obligations under the Agreement. A. GENERAL INFORMATION Party: United Kingdom Date of Report: July 2014 Period Covered by Report: January 2010 – December 2013 Competent Authority: The Department for Environment, Food and Rural Affairs (Defra) Changes Regarding: Competent Authority - Appointed member of the Advisory Committee - Membership of other committees/working groups - Abbreviations AHVLA Animal Health and Veterinary Laboratories Agency BCT Bat Conservation Trust CCW Countryside Council for Wales1 Defra Department for Environment, Food and Rural Affairs JNCC Joint Nature Conservation Committee GB Great Britain NBMP National Bat Monitoring Programme NE Natural England NERC Natural Environment Research Council NRW Natural Resources Wales SAC Special Area of Conservation SNH Scottish Natural Heritage SSI Site of Special Interest (Jersey classification) SSSI Site of Special Scientific Interest UK United Kingdom 1 With effect from 1 April 2013, CCW Environment Agency Wales and Forestry Commission Wales ceased to exist as their functions were transferred to a new body called Natural Resources Wales 1 B. STATUS OF BATS WITHIN THE TERRITORY OF THE PARTY 1. Summary Details of Resident Species UK Since the reported discovery of Alcathoe bat (Myotis alcathoe) at swarming sites in the UK, and then at swarming sites in Southern England (Sussex) in 2010, foraging specimens of the species have been reported to occur at a further site in Northern England (Yorkshire).