Gaits in Paramecium Escape
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Basal Body Structure and Composition in the Apicomplexans Toxoplasma and Plasmodium Maria E
Francia et al. Cilia (2016) 5:3 DOI 10.1186/s13630-016-0025-5 Cilia REVIEW Open Access Basal body structure and composition in the apicomplexans Toxoplasma and Plasmodium Maria E. Francia1* , Jean‑Francois Dubremetz2 and Naomi S. Morrissette3 Abstract The phylum Apicomplexa encompasses numerous important human and animal disease-causing parasites, includ‑ ing the Plasmodium species, and Toxoplasma gondii, causative agents of malaria and toxoplasmosis, respectively. Apicomplexans proliferate by asexual replication and can also undergo sexual recombination. Most life cycle stages of the parasite lack flagella; these structures only appear on male gametes. Although male gametes (microgametes) assemble a typical 9 2 axoneme, the structure of the templating basal body is poorly defined. Moreover, the rela‑ tionship between asexual+ stage centrioles and microgamete basal bodies remains unclear. While asexual stages of Plasmodium lack defined centriole structures, the asexual stages of Toxoplasma and closely related coccidian api‑ complexans contain centrioles that consist of nine singlet microtubules and a central tubule. There are relatively few ultra-structural images of Toxoplasma microgametes, which only develop in cat intestinal epithelium. Only a subset of these include sections through the basal body: to date, none have unambiguously captured organization of the basal body structure. Moreover, it is unclear whether this basal body is derived from pre-existing asexual stage centrioles or is synthesized de novo. Basal bodies in Plasmodium microgametes are thought to be synthesized de novo, and their assembly remains ill-defined. Apicomplexan genomes harbor genes encoding δ- and ε-tubulin homologs, potentially enabling these parasites to assemble a typical triplet basal body structure. -

Life: the Science of Biology
Lecture Notebook to accompany SINAUER ASSOCIATES, INC. MACMILLAN Copyright © 2014 Sinauer Associates, Inc. Cover photograph © Alex Mustard/naturepl.com. This document may not be modified or distributed (either electronically or on paper) without the permission of the publisher, with the following exception: Individual users may enter their own notes into this document and may print it for their own personal use. The Origin and Diversification of 0027 Eukaryotes 27.1 A Hypothetical Sequence for the Evolution of the Eukaryotic Cell (Page 550) 1 The protective Cell wall cell wall was lost. DNA 2 Infolding of the plasma membrane added surface area without increasing the cell’s volume. 3 Cytoskeleton (microfilament and microtubules) formed. 4 Internal membranes studded with ribosomes formed. 5 As regions of the infolded plasma membrane enclosed the cell’s DNA, a precursor of a nucleus formed. 6 Microtubules from the cytoskeleton formed the eukaryotic flagellum, 7 Early digestive vacuoles enabling propulsion. evolved into lysosomes using enzymes from the early endoplasmic reticulum. 8 Mitochondria formed through endosymbiosis with a proteobacterium. 9 Endosymbiosis with cyanobacteria led to the development of chloroplasts. Flagellum To add your own notes to any page, use Adobe Reader’s Chloroplast Typewriter feature, accessible via the Typewriter bar at Mitochondrion the top of the window. (Requires Adobe Reader 8 or later. Adobe Reader can be downloaded free of charge from the Nucleus Adobe website: http://get.adobe.com/reader.) LIFE2 The Science of Biology 10E Sadava © 2014 Sinauer Associates, Inc. Sinauer Associates Morales Studio Figure 27.01 05-24-12 Chapter 27 | The Origin and Diversification of Eukaryotes 3 (A) Primary endosymbiosis Eukaryote Cyanobacterium Cyanobacterium outer membrane Peptidoglycan Cyanobacterium inner membrane Host cell nucleus Chloroplast Peptidoglycan has been lost except in glaucophytes. -
Molecular Data and the Evolutionary History of Dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Un
Molecular data and the evolutionary history of dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Universitat Heidelberg, 1993 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES Department of Botany We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA November 2003 © Juan Fernando Saldarriaga Echavarria, 2003 ABSTRACT New sequences of ribosomal and protein genes were combined with available morphological and paleontological data to produce a phylogenetic framework for dinoflagellates. The evolutionary history of some of the major morphological features of the group was then investigated in the light of that framework. Phylogenetic trees of dinoflagellates based on the small subunit ribosomal RNA gene (SSU) are generally poorly resolved but include many well- supported clades, and while combined analyses of SSU and LSU (large subunit ribosomal RNA) improve the support for several nodes, they are still generally unsatisfactory. Protein-gene based trees lack the degree of species representation necessary for meaningful in-group phylogenetic analyses, but do provide important insights to the phylogenetic position of dinoflagellates as a whole and on the identity of their close relatives. Molecular data agree with paleontology in suggesting an early evolutionary radiation of the group, but whereas paleontological data include only taxa with fossilizable cysts, the new data examined here establish that this radiation event included all dinokaryotic lineages, including athecate forms. Plastids were lost and replaced many times in dinoflagellates, a situation entirely unique for this group. Histones could well have been lost earlier in the lineage than previously assumed. -

CH28 PROTISTS.Pptx
9/29/14 Biosc 41 Announcements 9/29 Review: History of Life v Quick review followed by lecture quiz (history & v How long ago is Earth thought to have formed? phylogeny) v What is thought to have been the first genetic material? v Lecture: Protists v Are we tetrapods? v Lab: Protozoa (animal-like protists) v Most atmospheric oxygen comes from photosynthesis v Lab exam 1 is Wed! (does not cover today’s lab) § Since many of the first organisms were photosynthetic (i.e. cyanobacteria), a LOT of excess oxygen accumulated (O2 revolution) § Some organisms adapted to use it (aerobic respiration) Review: History of Life Review: Phylogeny v Which organelles are thought to have originated as v Homology is similarity due to shared ancestry endosymbionts? v Analogy is similarity due to convergent evolution v During what event did fossils resembling modern taxa suddenly appear en masse? v A valid clade is monophyletic, meaning it consists of the ancestor taxon and all its descendants v How many mass extinctions seem to have occurred during v A paraphyletic grouping consists of an ancestral species and Earth’s history? Describe one? some, but not all, of the descendants v When is adaptive radiation likely to occur? v A polyphyletic grouping includes distantly related species but does not include their most recent common ancestor v Maximum parsimony assumes the tree requiring the fewest evolutionary events is most likely Quiz 3 (History and Phylogeny) BIOSC 041 1. How long ago is Earth thought to have formed? 2. Why might many organisms have evolved to use aerobic respiration? PROTISTS! Reference: Chapter 28 3. -

Tuberlatum Coatsi Gen. N., Sp. N. (Alveolata, Perkinsozoa), a New
Protist, Vol. 170, 82–103, February 2019 http://www.elsevier.de/protis Published online date 21 December 2018 ORIGINAL PAPER Tuberlatum coatsi gen. n., sp. n. (Alveolata, Perkinsozoa), a New Parasitoid with Short Germ Tubes Infecting Marine Dinoflagellates 1 Boo Seong Jeon, and Myung Gil Park LOHABE, Department of Oceanography, Chonnam National University, Gwangju 61186, Republic of Korea Submitted October 16, 2018; Accepted December 15, 2018 Monitoring Editor: Laure Guillou Perkinsozoa is an exclusively parasitic group within the alveolates and infections have been reported from various organisms, including marine shellfish, marine dinoflagellates, freshwater cryptophytes, and tadpoles. Despite its high abundance and great genetic diversity revealed by recent environmental rDNA sequencing studies, Perkinsozoa biodiversity remains poorly understood. During the intensive samplings in Korean coastal waters during June 2017, a new parasitoid of dinoflagellates was detected and was successfully established in culture. The new parasitoid was most characterized by the pres- ence of two to four dome-shaped, short germ tubes in the sporangium. The opened germ tubes were biconvex lens-shaped in the top view and were characterized by numerous wrinkles around their open- ings. Phylogenetic analyses based on the concatenated SSU and LSU rDNA sequences revealed that the new parasitoid was included in the family Parviluciferaceae, in which all members were comprised of two separate clades, one containing Parvilucifera species (P. infectans, P. corolla, and P. rostrata), and the other containing Dinovorax pyriformis, Snorkelia spp., and the new parasitoid from this study. Based on morphological, ultrastructural, and molecular data, we propose to erect a new genus and species, Tuberlatum coatsi gen. -

Lateral Gene Transfer of Anion-Conducting Channelrhodopsins Between Green Algae and Giant Viruses
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.15.042127; this version posted April 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 5 Lateral gene transfer of anion-conducting channelrhodopsins between green algae and giant viruses Andrey Rozenberg 1,5, Johannes Oppermann 2,5, Jonas Wietek 2,3, Rodrigo Gaston Fernandez Lahore 2, Ruth-Anne Sandaa 4, Gunnar Bratbak 4, Peter Hegemann 2,6, and Oded 10 Béjà 1,6 1Faculty of Biology, Technion - Israel Institute of Technology, Haifa 32000, Israel. 2Institute for Biology, Experimental Biophysics, Humboldt-Universität zu Berlin, Invalidenstraße 42, Berlin 10115, Germany. 3Present address: Department of Neurobiology, Weizmann 15 Institute of Science, Rehovot 7610001, Israel. 4Department of Biological Sciences, University of Bergen, N-5020 Bergen, Norway. 5These authors contributed equally: Andrey Rozenberg, Johannes Oppermann. 6These authors jointly supervised this work: Peter Hegemann, Oded Béjà. e-mail: [email protected] ; [email protected] 20 ABSTRACT Channelrhodopsins (ChRs) are algal light-gated ion channels widely used as optogenetic tools for manipulating neuronal activity 1,2. Four ChR families are currently known. Green algal 3–5 and cryptophyte 6 cation-conducting ChRs (CCRs), cryptophyte anion-conducting ChRs (ACRs) 7, and the MerMAID ChRs 8. Here we 25 report the discovery of a new family of phylogenetically distinct ChRs encoded by marine giant viruses and acquired from their unicellular green algal prasinophyte hosts. -

Chloroplast Structure of the Cryptophyceae
CHLOROPLAST STRUCTURE OF THE CRYPTOPHYCEAE Evidence for Phycobiliproteins within Intrathylakoidal Spaces E . GANTT, M . R . EDWARDS, and L . PROVASOLI From the Radiation Biology Laboratory, Smithsonian Institution, Rockville, Maryland 20852, the Division of Laboratories and Research, New York State Department of Health, Albany, New York 12201, and the Haskins Laboratories, New Haven, Connecticut 06520 ABSTRACT Selective extraction and morphological evidence indicate that the phycobiliproteins in three Cryptophyceaen algae (Chroomonas, Rhodomonas, and Cryptomonas) are contained within intrathylakoidal spaces and are not on the stromal side of the lamellae as in the red and blue-green algae . Furthermore, no discrete phycobilisome-type aggregates have thus far been observed in the Cryptophyceae . Structurally, although not necessarily functionally, this is a radical difference . The width of the intrathylakoidal spaces can vary but is gen- erally about 200-300 A . While the thylakoid membranes are usually closely aligned, grana- type fusions do not occur. In Chroomonas these membranes evidence an extensive periodic display with a spacing on the order of 140-160 A . This periodicity is restricted to the mem- branes and has not been observed in the electron-opaque intrathylakoidal matrix . INTRODUCTION The varied characteristics of the cryptomonads (3), Greenwood in Kirk and Tilney-Bassett (10), are responsible for their indefinite taxonomic and Lucas (13) . The thylakoids have a tendency position (17), but at the same time they enhance to be arranged in pairs, that is, for two of them to their status in evolutionary schemes (1, 4) . Their be closely associated with a 30-50 A space between chloroplast structure is distinct from that of every them . -

Dinophyceae), with Notes on Feeding Behavior and the Description of the Flagellar Base Area of a Planozygote1
J. Phycol. 42, 434–452 (2006) r 2006 Phycological Society of America DOI: 10.1111/j.1529-8817.2006.00195.x ULTRASTRUCTURE AND LSU rDNA-BASED PHYLOGENY OF ESOPTRODINIUM GEMMA (DINOPHYCEAE), WITH NOTES ON FEEDING BEHAVIOR AND THE DESCRIPTION OF THE FLAGELLAR BASE AREA OF A PLANOZYGOTE1 Anto´nio J. Calado,2 Sandra C. Craveiro Departamento de Biologia, Universidade de Aveiro, P-3810-193 Aveiro, Portugal Niels Daugbjerg and Øjvind Moestrup Department of Phycology, Biological Institute, University of Copenhagen, Øster Farimagsgade 2D, DK-1353 Copenhagen K, Denmark A small, freshwater dinoflagellate with an incom- Markov chains; MP, maximum parsimony; NJ, plete cingulum, identified as Esoptrodinium gemma neighbor-joining; PP, posterior probabilities; PSC, Javornicky´ (5Bernardinium bernardinense sensu au- peduncular striated collar; SRC, striated root con- ctt. non sensu Chodat), was maintained in mixed nective (TSR to LMR); TB, transverse basal body; culture and examined using light and serial section TMR, transverse microtubular root; TMRE, TMR TEM. Vegetative flagellate cells, large cells with two extension; TSR, transverse striated root; TSRM, longitudinal flagella (planozygotes), and cysts were transverse striated root microtubule; VR, ventral examined. The cells displayed a red eyespot near ridge the base of the longitudinal flagellum, made of two or three layers of pigment globules not bounded by a membrane. Yellow-green, band-shaped chloro- plasts, bounded by three membranes and contain- The organism used in this study is a naked dino- ing lamella with three thylakoids, were present in flagellate that would be readily identified as Bern- both flagellate cells and cysts. Most cells had food ardinium bernardinense Chodat by recent dinoflagellate vacuoles, containing phagotrophically ingested floras (Starmach 1974, Matvienko and Litvinenko chlamydomonads or chlorelloid green algae; inges- 1977, Popovsky´ and Pfiester 1990). -

Download File
Acta Protozool. (2012) 51: 305–318 http://www.eko.uj.edu.pl/ap ACTA doi:10.4467/16890027AP.12.024.0784 PROTOZOOLOGICA Morphological Description of Telaepolella tubasferens n. g. n. sp., Isolate ATCC© 50593™, a Filose Amoeba in the Gracilipodida, Amoebozoa Daniel J. G. LAHR1,2*, Gabriela M. KUBIK1*, Anastasia L. GANT1, Jessica GRANT1, O. Roger ANDERSON3 and Laura A. KATZ1,2 1Department of Biological Sciences, Smith College, Northampton, MA, USA; 2Program in Organismic and Evolutionary Biology, University of Massachusetts, Amherst, MA, USA; 3Biology, Lamont-Doherty Earth Observatory of Columbia University, Palisades, New York; * D. J. G. Lahr and G. M. Kubik contributed equally to this work Abstract. We describe the amoeboid isolate ATCC© 50593™ as a new taxon, Telaepolella tubasferens n. g. n. sp. This multinucleated amoeba has filose pseudopods and is superficially similar to members of the vampyrellids (Rhizaria) such as Arachnula impatiens Cien- kowski, 1876, which was the original identification upon deposition. However, previous multigene analyses place this taxon within the Gracilipodida Lahr and Katz 2011 in the Amoebozoa. Here, we document the morphology of this organism at multiple life history stages and provide data underlying the description as a new taxon. We demonstrate that T. tubasferens is distinct from Arachnula and other rhizari- ans (Theratromyxa, Leptophrys) in a suite of morphological characters such as general body shape, relative size of pseudopods, distinction of ecto- and endoplasmic regions, and visibility of nuclei in non-stained cells (an important diagnostic character). Although Amoebozoa taxa generally have lobose pseudopods, genera in Gracilipodida such as Flamella and Filamoeba as well as several organisms previously classified as protosteloid amoebae (e.g. -
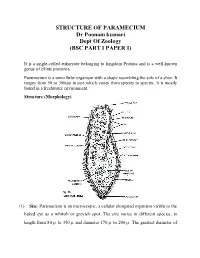
STRUCTURE of PARAMECIUM Dr Poonam Kumari Dept of Zoology (BSC PART I PAPER I)
STRUCTURE OF PARAMECIUM Dr Poonam kumari Dept Of Zoology (BSC PART I PAPER I) It is a single-celled eukaryote belonging to kingdom Protista and is a well-known genus of ciliate protozoa. Paramecium is a unicellular organism with a shape resembling the sole of a shoe. It ranges from 50 to 300um in size which varies from species to species. It is mostly found in a freshwater environment. Structure (Morphology) (1) Size: Paramecium is an microscopic, a cellular elongated organism visible to the baked eye as a whitish or greyish spot. The size varies in different species, in length from 80 to 350 and diameter 170 to 290 . The greatest diameter of the cylindrical body is about two third of its entire length. Usually the individuals of the same species may show minor morphological and physiological differences. (2) Shape: Paramecium is a slipper shaped, cigar shaped, or spindle shaped animalcule. Its shape is usually constant and a symmetrical, because slipper like shape. The body is elongated, blunt and rounded at the anterior end and somewhat pointed of the posterior end. In cross section it is circular with greatest diameter behind the centre of body. The anterior half of the body is slightly twisted. The body is distinguished into an oral or ventral surface and an aboral or dorsal surface. The structure is more complicated due to the development of certain organelles in the acellular body. (3) Oral groove: The ventral surface of body bears a prominent, oblique and shallow depression is called oral groove, it arise from the middle of body and extends to the left side of anterior end. -

Giant Protistan Parasites on the Gills of Cephalopods (Mollusca)
DISEASES OF AQUATIC ORGANISMS Vol. 3: 119-125. 1987 Published December 14 Dis. aquat. Org. Giant protistan parasites on the gills of cephalopods (Mollusca) Norman ~c~ean',F. G. ~ochberg~,George L. shinn3 ' Biology Department, San Diego State University, San Diego, California 92182-0057, USA Department of Invertebrate Zoology, Santa Barbara Museum of Natural History, 2559 Puesta Del Sol Road, Santa Barbara, California 93105. USA Division of Science, Northeast Missouri State University, Kirksville. Missouri 63501, USA ABSTRACT: Large Protista of unknown taxonomic affinities are described from 3 species of coleoid squids, and are reported from many other species of cephalopods. The white to yellow-orange, ovoid cyst-like parasites are partially embedded within small pockets on the surface of the gills, often in large numbers. Except for a holdfast region on one side of the large end, the surface of the parasite is elaborated into low triangular plates separated by grooves. The parasites are uninucleate; their cytoplasm bears lipid droplets and presumed paraglycogen granules. Trichocysts, present in a layer beneath the cytoplasmic surface, were found by transmission electron microscopy to be of the dino- flagellate type. Further studies are needed to clarify the taxonomic position of these protists. INTRODUCTION epoxy resin (see below). One specimen each of the coleoid squids Abralia trigonura and Histioteuthis dof- Cephalopods harbor a diversity of metazoan and leini were trawled near Oahu, Hawaii, in March, 1980. protozoan parasites (Hochberg 1983). In this study we Gill parasites from the former were fixed in formalin; used light and electron microscopy to characterize a those from the latter were fixed in osmium tetroxide. -

Diversity of Life Paramecia—Paramecium Caudatum
Used in: Diversity of Life Paramecia—Paramecium caudatum Background. Paramecia are single-celled ciliated protists found in freshwater ponds. They feed on microorganisms such as bacteria, algae, and yeasts, sweeping the food down the oral groove, into the mouth. Their movement is characterized by whiplike movement of the cilia, small hair-like projections that are arranged along the outside of their bodies. They spiral through the water until running into an obstacle, at which point the cilia "reverse course" so the paramecium can swim backwards and try again. Paramecia have two nuclei and reproduce asexually, by binary fission. A paramecium can also exchange genetic material with another via the process of conjugation. Acquiring paramecia. You can purchase Paramecium caudatum from Delta Education or a biological supply house. This species is a classic classroom organism, hardy and large enough for students to easily observe using a light microscope. Purchase enough to "spike" a sample of water that students will use for preparing slides of elodea leaves and to use in Part 2 of Investigation 3 when students will focus specifically on study of the organism itself. What to do when they arrive. Open the shipping container, remove the culture jar, and loosen the lid on the jar. Aerate the culture using the pipette supplied, bubbling air through the water. Repeat several times to oxygenate the water. After about 15 minutes, use a dropper or the pipette to obtain organisms, gathering them from around the barley (or other food source). Prepare a wet-mount slide and look for paramecia using a microscope.