APOBEC4, a New Member of the AID/APOBEC Family of Polynucleotide (Deoxy)Cytidine Deaminases Predicted by Computational Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Antibody List
產品編號 產品名稱 PA569955 1110059E24Rik Polyclonal Antibody PA569956 1110059E24Rik Polyclonal Antibody PA570131 1190002N15Rik Polyclonal Antibody 01-1234-42 123count eBeads Counting Beads MA512242 14.3.3 Pan Monoclonal Antibody (CG15) LFMA0074 14-3-3 beta Monoclonal Antibody (60C10) LFPA0077 14-3-3 beta Polyclonal Antibody PA137002 14-3-3 beta Polyclonal Antibody PA14647 14-3-3 beta Polyclonal Antibody PA515477 14-3-3 beta Polyclonal Antibody PA517425 14-3-3 beta Polyclonal Antibody PA522264 14-3-3 beta Polyclonal Antibody PA529689 14-3-3 beta Polyclonal Antibody MA134561 14-3-3 beta/epsilon/zeta Monoclonal Antibody (3C8) MA125492 14-3-3 beta/zeta Monoclonal Antibody (22-IID8B) MA125665 14-3-3 beta/zeta Monoclonal Antibody (4E2) 702477 14-3-3 delta/zeta Antibody (1H9L19), ABfinity Rabbit Monoclonal 711507 14-3-3 delta/zeta Antibody (1HCLC), ABfinity Rabbit Oligoclonal 702241 14-3-3 epsilon Antibody (5H10L5), ABfinity Rabbit Monoclonal 711273 14-3-3 epsilon Antibody (5HCLC), ABfinity Rabbit Oligoclonal PA517104 14-3-3 epsilon Polyclonal Antibody PA528937 14-3-3 epsilon Polyclonal Antibody PA529773 14-3-3 epsilon Polyclonal Antibody PA575298 14-3-3 eta (Lys81) Polyclonal Antibody MA524792 14-3-3 eta Monoclonal Antibody PA528113 14-3-3 eta Polyclonal Antibody PA529774 14-3-3 eta Polyclonal Antibody PA546811 14-3-3 eta Polyclonal Antibody MA116588 14-3-3 gamma Monoclonal Antibody (HS23) MA116587 14-3-3 gamma Monoclonal Antibody (KC21) PA529690 14-3-3 gamma Polyclonal Antibody PA578233 14-3-3 gamma Polyclonal Antibody 510700 14-3-3 Pan Polyclonal -

Download Download
Robyn A Lindley. Medical Research Archives vol 8 issue 8. Medical Research Archives REVIEW ARTICLE Review of the mutational role of deaminases and the generation of a cognate molecular model to explain cancer mutation spectra Author Robyn A Lindley1,2 1Department of Clinical Pathology 2GMDx Genomics Ltd, The Victorian Comprehensive Cancer Centre Level 3 162 Collins Street, Faculty of Medicine, Dentistry & Health Sciences Melbourne VIC3000, AUSTRALIA University of Melbourne, Email: [email protected] 305 Gratton Street, Melbourne, VIC 3000, AUSTRALIA Email: [email protected] Correspondence: Robyn A Lindley, Department of Clinical Pathology, Faculty of Medicine, Dentistry & Health Sciences, University of Melbourne, 305 Gratton Street, Melbourne VIC 3000 AUSTRALIA Mobile: +61 (0) 414209132 Email: [email protected] Abstract Recent developments in somatic mutation analyses have led to the discovery of codon-context targeted somatic mutation (TSM) signatures in cancer genomes: it is now known that deaminase mutation target sites are far more specific than previously thought. As this research provides novel insights into the deaminase origin of most of the somatic point mutations arising in cancer, a clear understanding of the mechanisms and processes involved will be valuable for molecular scientists as well as oncologists and cancer specialists in the clinic. This review will describe the basic research into the mechanism of antigen-driven somatic hypermutation of immunoglobulin variable genes (Ig SHM) that lead to the discovery of TSM signatures, and it will show that an Ig SHM-like signature is ubiquitous in the cancer exome. Most importantly, the data discussed in this review show that Ig SHM-like cancer-associated signatures are highly targeted to cytosine (C) and adenosine (A) nucleotides in a characteristic codon-context fashion. -

WO2019226953A1.Pdf
) ( 2 (51) International Patent Classification: Street, Brookline, MA 02446 (US). WILSON, Christo¬ C12N 9/22 (2006.01) pher, Gerard; 696 Main Street, Apartment 311, Waltham, MA 0245 1(US). DOMAN, Jordan, Leigh; 25 Avon Street, (21) International Application Number: Somverville, MA 02143 (US). PCT/US20 19/033 848 (74) Agent: HEBERT, Alan, M. et al. ;Wolf, Greenfield, Sacks, (22) International Filing Date: P.C., 600 Atlanitc Avenue, Boston, MA 02210-2206 (US). 23 May 2019 (23.05.2019) (81) Designated States (unless otherwise indicated, for every (25) Filing Language: English kind of national protection av ailable) . AE, AG, AL, AM, (26) Publication Language: English AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (30) Priority Data: DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, 62/675,726 23 May 2018 (23.05.2018) US HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, 62/677,658 29 May 2018 (29.05.2018) US KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (71) Applicants: THE BROAD INSTITUTE, INC. [US/US]; MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, 415 Main Street, Cambridge, MA 02142 (US). PRESI¬ OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, DENT AND FELLOWS OF HARVARD COLLEGE SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, [US/US]; 17 Quincy Street, Cambridge, MA 02138 (US). -

Genesymbol Bindingsite Motifs Pvalue Description Ensembl Gene Id AASDH Chr4:57207615-57208306 1 9.55E-79
GeneSymbol BindingSite Motifs pValue description ensembl_gene_id AASDH chr4:57207615-57208306 1 9.55E-79 aminoadipate-semialdehyde dehydrogenase ENSG00000157426 ABCA10 chr17:67259657-67260436 1 2.57E-85 ATP-binding cassette, sub-family A (ABC1), member 10 ENSG00000154263 ABCB9 chr12:123415801-123416656 1 5.89E-77 ATP-binding cassette, sub-family B (MDR/TAP), member 9 ENSG00000150967 ABCG5 chr2:44039916-44040709 1 1.66E-75 ATP-binding cassette, sub-family G (WHITE), member 5 ENSG00000138075 ABCG8 chr2:44136156-44136727 1 5.13E-87 ATP-binding cassette, sub-family G (WHITE), member 8 ENSG00000143921 ABHD5 chr3:43717538-43718405 1 9.77E-76 abhydrolase domain containing 5 ENSG00000011198 ABL2 chr1:179167916-179168697 1 3.16E-87 c-abl oncogene 2, non-receptor tyrosine kinase ENSG00000143322 ABRA chr8:107672555-107673551 1 1.48E-107 actin-binding Rho activating protein ENSG00000174429 ACADL chr2:211076683-211077481 1 2.40E-77 acyl-CoA dehydrogenase, long chain ENSG00000115361 ACADM chr1:76190984-76192200 1 2.40E-135 acyl-CoA dehydrogenase, C-4 to C-12 straight chain ENSG00000117054 ACAT1 chr11:107996439-107997181 1 2.09E-76 acetyl-CoA acetyltransferase 1 ENSG00000075239 ACP6 chr1:147102212-147103195 1 1.55E-80 acid phosphatase 6, lysophosphatidic ENSG00000162836 ACSL4 chrX:109046263-109047211 1 4.79E-84 acyl-CoA synthetase long-chain family member 4 ENSG00000068366 ACSM4 chr12:7419079-7419616 1 3.72E-72 acyl-CoA synthetase medium-chain family member 4 ENSG00000215009 ACTRT1 chrX:126916430-127482343 4 2.01E-78 actin-related protein T1 ENSG00000123165 -

Supplemental Table S1. Primers for Sybrgreen Quantitative RT-PCR Assays
Supplemental Table S1. Primers for SYBRGreen quantitative RT-PCR assays. Gene Accession Primer Sequence Length Start Stop Tm GC% GAPDH NM_002046.3 GAPDH F TCCTGTTCGACAGTCAGCCGCA 22 39 60 60.43 59.09 GAPDH R GCGCCCAATACGACCAAATCCGT 23 150 128 60.12 56.52 Exon junction 131/132 (reverse primer) on template NM_002046.3 DNAH6 NM_001370.1 DNAH6 F GGGCCTGGTGCTGCTTTGATGA 22 4690 4711 59.66 59.09% DNAH6 R TAGAGAGCTTTGCCGCTTTGGCG 23 4797 4775 60.06 56.52% Exon junction 4790/4791 (reverse primer) on template NM_001370.1 DNAH7 NM_018897.2 DNAH7 F TGCTGCATGAGCGGGCGATTA 21 9973 9993 59.25 57.14% DNAH7 R AGGAAGCCATGTACAAAGGTTGGCA 25 10073 10049 58.85 48.00% Exon junction 9989/9990 (forward primer) on template NM_018897.2 DNAI1 NM_012144.2 DNAI1 F AACAGATGTGCCTGCAGCTGGG 22 673 694 59.67 59.09 DNAI1 R TCTCGATCCCGGACAGGGTTGT 22 822 801 59.07 59.09 Exon junction 814/815 (reverse primer) on template NM_012144.2 RPGRIP1L NM_015272.2 RPGRIP1L F TCCCAAGGTTTCACAAGAAGGCAGT 25 3118 3142 58.5 48.00% RPGRIP1L R TGCCAAGCTTTGTTCTGCAAGCTGA 25 3238 3214 60.06 48.00% Exon junction 3124/3125 (forward primer) on template NM_015272.2 Supplemental Table S2. Transcripts that differentiate IPF/UIP from controls at 5%FDR Fold- p-value Change Transcript Gene p-value p-value p-value (IPF/UIP (IPF/UIP Cluster ID RefSeq Symbol gene_assignment (Age) (Gender) (Smoking) vs. C) vs. C) NM_001178008 // CBS // cystathionine-beta- 8070632 NM_001178008 CBS synthase // 21q22.3 // 875 /// NM_0000 0.456642 0.314761 0.418564 4.83E-36 -2.23 NM_003013 // SFRP2 // secreted frizzled- 8103254 NM_003013 -

Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins
microorganisms Review Deaminase-Independent Mode of Antiretroviral Action in Human and Mouse APOBEC3 Proteins Yoshiyuki Hakata 1,* and Masaaki Miyazawa 1,2 1 Department of Immunology, Kindai University Faculty of Medicine, 377-2 Ohno-Higashi, Osaka-Sayama, Osaka 589-8511, Japan; [email protected] 2 Kindai University Anti-Aging Center, 3-4-1 Kowakae, Higashiosaka, Osaka 577-8502, Japan * Correspondence: [email protected]; Tel.: +81-72-367-7660 Received: 8 December 2020; Accepted: 9 December 2020; Published: 12 December 2020 Abstract: Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3 (APOBEC3) proteins (APOBEC3s) are deaminases that convert cytosines to uracils predominantly on a single-stranded DNA, and function as intrinsic restriction factors in the innate immune system to suppress replication of viruses (including retroviruses) and movement of retrotransposons. Enzymatic activity is supposed to be essential for the APOBEC3 antiviral function. However, it is not the only way that APOBEC3s exert their biological function. Since the discovery of human APOBEC3G as a restriction factor for HIV-1, the deaminase-independent mode of action has been observed. At present, it is apparent that both the deaminase-dependent and -independent pathways are tightly involved not only in combating viruses but also in human tumorigenesis. Although the deaminase-dependent pathway has been extensively characterized so far, understanding of the deaminase-independent pathway remains immature. Here, we review existing knowledge regarding the deaminase-independent antiretroviral functions of APOBEC3s and their molecular mechanisms. We also discuss the possible unidentified molecular mechanism for the deaminase-independent antiretroviral function mediated by mouse APOBEC3. Keywords: APOBEC3; deaminase-independent antiretroviral function; innate immunity 1. -

Diversification of AID/APOBEC-Like Deaminases in Metazoa: Multiplicity
Diversification of AID/APOBEC-like deaminases in PNAS PLUS metazoa: multiplicity of clades and widespread roles in immunity Arunkumar Krishnana, Lakshminarayan M. Iyera, Stephen J. Hollandb, Thomas Boehmb, and L. Aravinda,1 aNational Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894; and bDepartment of Developmental Immunology, Max Planck Institute of Immunobiology and Epigenetics, 79108 Freiburg, Germany Edited by Anjana Rao, La Jolla Institute and University of California San Diego, La Jolla, CA, and approved February 23, 2018 (received for review November 30, 2017) AID/APOBEC deaminases (AADs) convert cytidine to uridine in viruses and retrotransposons through hypermutation of DNA during single-stranded nucleic acids. They are involved in numerous muta- reverse transcription (APOBEC3s) (21). genic processes, including those underpinning vertebrate innate and The deaminase superfamily displays a conserved β-sheet with adaptive immunity. Using a multipronged sequence analysis strategy, five β-strands arranged in 2-1-3-4-5 order interleaved with three we uncover several AADs across metazoa, dictyosteliida, and algae, α-helices forming an α/β-fold (the deaminase fold) (22), which it including multiple previously unreported vertebrate clades, and shares with JAB/RadC, AICAR transformylase, formate de- versions from urochordates, nematodes, echinoderms, arthropods, hydrogenase accessory subunit (FdhD), and Tm1506 superfamilies lophotrochozoans, cnidarians, and porifera. Evolutionary analysis sug- of proteins. The active site consists of two zinc (Zn)-chelating gests a fundamental division of AADs early in metazoan evolution into motifs, respectively typified by the signatures HxE/CxE/DxE at secreted deaminases (SNADs) and classical AADs, followed by diver- the end of helix 2 and CxnC (where x is any amino acid and n is ≥2) sification into several clades driven by rapid-sequence evolution, gene located in loop 5 and the beginning of helix 3 (Fig. -

Insights Into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions
viruses Review Insights into the Structures and Multimeric Status of APOBEC Proteins Involved in Viral Restriction and Other Cellular Functions Xiaojiang S. Chen 1,2,3 1 Molecular and Computational Biology, Departments of Biological Sciences, Chemistry, University of Southern California, Los Angeles, CA 90089, USA; [email protected]; Tel.: +1-213-740-5487 2 Genetic, Molecular and Cellular Biology Program, Keck School of Medicine, Norris Comprehensive Cancer Center, University of Southern California, Los Angeles, CA 90089, USA 3 Center of Excellence in NanoBiophysics/Structural Biology, University of Southern California, Los Angeles, CA 90089, USA Abstract: Apolipoprotein B mRNA editing catalytic polypeptide-like (APOBEC) proteins belong to a family of deaminase proteins that can catalyze the deamination of cytosine to uracil on single- stranded DNA or/and RNA. APOBEC proteins are involved in diverse biological functions, including adaptive and innate immunity, which are critical for restricting viral infection and endogenous retroelements. Dysregulation of their functions can cause undesired genomic mutations and RNA modification, leading to various associated diseases, such as hyper-IgM syndrome and cancer. This review focuses on the structural and biochemical data on the multimerization status of individual APOBECs and the associated functional implications. Many APOBECs form various multimeric complexes, and multimerization is an important way to regulate functions for some of these proteins at several levels, such as deaminase activity, protein stability, subcellular localization, protein storage Citation: Chen, X.S. Insights into and activation, virion packaging, and antiviral activity. The multimerization of some APOBECs is the Structures and Multimeric Status more complicated than others, due to the associated complex RNA binding modes. -
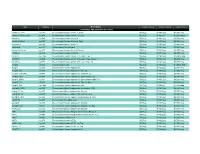
Prospec (Product
Name Catalog # Description Customer Price-A Customer Price-B Customer Price-C CYTOKINES AND GROWTH FACTORS Activin A, Active cyt-145 Recombinant Human Activin-A, Active $50/2µg $130/10µg $3,500/1mg Activin A, Plant-Active cyt-414 Recombinant Human Activin-A Active $50/1µg $130/5µg $1,500/100µg Activin A cyt-569 Recombinant Human Activin-A $50/2µg $130/10µg $2,700/1mg Activin A, Plant cyt-052 Recombinant Human Activin-A, Plant $50/2µg $130/10µg $4,800/1mg mActivin A cyt-146 Recombinant Mouse Activin-A $50/2µg $130/10µg $3,500/1mg rActivin A cyt-147 Recombinant Rat Activin-A $50/2µg $130/10µg $3,500/1mg Activin B, Active cyt-057 Recombinant Human Activin-B Active $50/2µg $130/10µg $5,200/1mg Activin B cyt-058 Recombinant Human Activin-B $50/2µg $130/10µg $4,800/1mg ACVR1 cyt-1140 Recombinant Human Activin A Receptor Type 1 $50/2µg $130/10µg $1,000/0.1mg ACVRL1 cyt-920 Recombinant Human Activin A Receptor Type II-Like 1 $50/2µg $130/10µg $5,200/1mg ACVR2A cyt-976 Recombinant Human Activin A Receptor Type 2A $50/2µg $130/10µg $5,200/1mg Acrp30 cyt-024 Human Adiponectin $50/2µg $130/10µg $1,000/0.1mg Acrp30 cyt-280 Recombinant Human Adiponectin $50/5µg $130/25µg $2,700/1mg Acrp30, His cyt-433 Recombinant Human Adiponectin, His tag $50/10µg $130/50µg $1,900/1mg Acrp30 (108-244) cyt-073 Recombinant Human Adiponectin (108-244 a.a.) $50/5µg $130/25µg $2,700/1mg Acrp30, HEK cyt-434 Recombinant Human Adiponectin glycosilated, HEK $50/2µg $130/10µg $4,680/1mg Acrp30, HMW cyt-764 Recombinant Human Adiponectin glycosilated, HMW Rich $50/2µg $130/10µg -

Prospec Product Catalog 2019
ProSpec Product Catalog 2019 QUALITY PROTEINS for scientific discoveries WE'VE GOT CHEMISTRY BRIGHTEN YOUR RESEARCH part for life sciences 6,000 proteins at your fingertips List by Molecule Name List by Molecule Name 제품별 자세한 설명은 www.prospecbio.com을 방문하세요. 부가세 별도 Molecule 제품번호 제품명 Size A / 가격(원) Size B / 가격(원) Size C / 가격(원) Name 1F8 Chagas cch-002 Recombinant 1F8 Chagas 5µg / 90,500 20µg / 199,000 1mg / 5,110,500 4-1BBL cyt-649 Recombinant Human 4-1BB Ligand, His Tag 5µg / 112,200 20µg / 220,800 1mg / 3,793,200 4-1BBL cyt-149 Recombinant Human 4-1BB Ligand 5µg / 90,500 20µg / 199,000 1mg / 3,771,400 4-1BBR cyt-916 Recombinant Mouse 4-1BB Receptor 1µg / 112,200 5µg / 220,800 50µg / 1,701,000 4-1BBR cyt-463 Recombinant Human 4-1BB Receptor 5µg / 90,500 20µg / 199,000 1mg / 3,771,400 4-1BBR cyt-137 Recombinant Human 4-1BB Receptor His Tag 5µg / 112,200 20µg / 220,800 1mg / 5,016,800 4-1BBR cyt-931 Recombinant Human 4-1BB Receptor sf9 2µg / 112,200 10µg / 220,800 1mg / 7,227,300 A2LD1 pro-172 Recombinant Human AIG2-Like Domain 1 2µg / 112,200 10µg / 220,800 1mg / 7,227,300 A2M pro-551 Human Alpha-2 Macroglobulin Protein 200µg / 90,500 1mg / 317,500 10mg / 2,389,800 Recombinant Human Alpha & Gamma-Adaptin Binding AAGAB pro-1479 5µg / 112,200 20µg / 220,800 1mg / 3,793,200 Protein Recombinant Human Adipogenesis Associated, Mth938 AAMDC pro-2116 5µg / 112,200 20µg / 220,800 1mg / 3,793,200 Domain Containing AARS enz-305 Recombinant Human Alanyl t-RNA Synthetase 5µg / 112,200 20µg / 220,800 1mg / 5,372,000 Recombinant Human Aminoadipate-Semialdehyde -

Supplemental Materials
The infection-tolerant mammalian reservoir of Lyme disease and other zoonoses broadly counters the inflammatory effects of endotoxin Supplemental Materials Figures S1-S5 Tables S1-S20 Figure S1. Digital photograph of two adult Peromyscus leucopus with exudative conjunctivitis and huddled together. The animals had received 10 mg/gm body of Escherichia coli lipopolysaccharide intraperitoneally the day before. Figure S2. Species- and tissue-specific responses to LPS. Independent differential gene expression analysis of RNA-seq data were performed for blood, spleen, and liver tissues of P. leucopus and M. musculus collected 4 h after injection with LPS or buffer alsone as control. These are represented as volcano plots with range- adjusted scales for the log2-transformed fold-changes on x-axes and log10-transformed FDR p values on y- axes. Colors of symbols denote the following: red, up-regulated gene with absolute fold-change > 4.0 and p value < 0.05; purple, down-regulated gene with absolute fold-change > 4.0 and p value < 0.05; and gray, all others. Numbers at the top left and right corners in each plot respresent numbers of down- and up-regulated genes, respectively. Figure 3 is same data with constant scales for x- and y-axes across the plots. Numerical values for each gene in the 6 datasets are provided in Tables S4-S9. Figure S3. Correlation of IL-10 and IL-10 P. leucopus and M. musculus from RNA-seq of spleen and of Figure 6B and TaBle S14. The scatter plot is log10 values of normalized unique reads of one coding sequence against another for each of the four groups, as defined in the legend for Figure 6 and indicated By different symBols.