(Bio-)Chemical Tools and Visualization Software for Computational Science
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

GPAW, Gpus, and LUMI
GPAW, GPUs, and LUMI Martti Louhivuori, CSC - IT Center for Science Jussi Enkovaara GPAW 2021: Users and Developers Meeting, 2021-06-01 Outline LUMI supercomputer Brief history of GPAW with GPUs GPUs and DFT Current status Roadmap LUMI - EuroHPC system of the North Pre-exascale system with AMD CPUs and GPUs ~ 550 Pflop/s performance Half of the resources dedicated to consortium members Programming for LUMI Finland, Belgium, Czechia, MPI between nodes / GPUs Denmark, Estonia, Iceland, HIP and OpenMP for GPUs Norway, Poland, Sweden, and how to use Python with AMD Switzerland GPUs? https://www.lumi-supercomputer.eu GPAW and GPUs: history (1/2) Early proof-of-concept implementation for NVIDIA GPUs in 2012 ground state DFT and real-time TD-DFT with finite-difference basis separate version for RPA with plane-waves Hakala et al. in "Electronic Structure Calculations on Graphics Processing Units", Wiley (2016), https://doi.org/10.1002/9781118670712 PyCUDA, cuBLAS, cuFFT, custom CUDA kernels Promising performance with factor of 4-8 speedup in best cases (CPU node vs. GPU node) GPAW and GPUs: history (2/2) Code base diverged from the main branch quite a bit proof-of-concept implementation had lots of quick and dirty hacks fixes and features were pulled from other branches and patches no proper unit tests for GPU functionality active development stopped soon after publications Before development re-started, code didn't even work anymore on modern GPUs without applying a few small patches Lesson learned: try to always get new functionality to the -

MODELLER 10.1 Manual
MODELLER A Program for Protein Structure Modeling Release 10.1, r12156 Andrej Saliˇ with help from Ben Webb, M.S. Madhusudhan, Min-Yi Shen, Guangqiang Dong, Marc A. Martı-Renom, Narayanan Eswar, Frank Alber, Maya Topf, Baldomero Oliva, Andr´as Fiser, Roberto S´anchez, Bozidar Yerkovich, Azat Badretdinov, Francisco Melo, John P. Overington, and Eric Feyfant email: modeller-care AT salilab.org URL https://salilab.org/modeller/ 2021/03/12 ii Contents Copyright notice xxi Acknowledgments xxv 1 Introduction 1 1.1 What is Modeller?............................................. 1 1.2 Modeller bibliography....................................... .... 2 1.3 Obtainingandinstallingtheprogram. .................... 3 1.4 Bugreports...................................... ............ 4 1.5 Method for comparative protein structure modeling by Modeller ................... 5 1.6 Using Modeller forcomparativemodeling. ... 8 1.6.1 Preparinginputfiles . ............. 8 1.6.2 Running Modeller ......................................... 9 2 Automated comparative modeling with AutoModel 11 2.1 Simpleusage ..................................... ............ 11 2.2 Moreadvancedusage............................... .............. 12 2.2.1 Including water molecules, HETATM residues, and hydrogenatoms .............. 12 2.2.2 Changing the default optimization and refinement protocol ................... 14 2.2.3 Getting a very fast and approximate model . ................. 14 2.2.4 Building a model from multiple templates . .................. 15 2.2.5 Buildinganallhydrogenmodel -

Free and Open Source Software for Computational Chemistry Education
Free and Open Source Software for Computational Chemistry Education Susi Lehtola∗,y and Antti J. Karttunenz yMolecular Sciences Software Institute, Blacksburg, Virginia 24061, United States zDepartment of Chemistry and Materials Science, Aalto University, Espoo, Finland E-mail: [email protected].fi Abstract Long in the making, computational chemistry for the masses [J. Chem. Educ. 1996, 73, 104] is finally here. We point out the existence of a variety of free and open source software (FOSS) packages for computational chemistry that offer a wide range of functionality all the way from approximate semiempirical calculations with tight- binding density functional theory to sophisticated ab initio wave function methods such as coupled-cluster theory, both for molecular and for solid-state systems. By their very definition, FOSS packages allow usage for whatever purpose by anyone, meaning they can also be used in industrial applications without limitation. Also, FOSS software has no limitations to redistribution in source or binary form, allowing their easy distribution and installation by third parties. Many FOSS scientific software packages are available as part of popular Linux distributions, and other package managers such as pip and conda. Combined with the remarkable increase in the power of personal devices—which rival that of the fastest supercomputers in the world of the 1990s—a decentralized model for teaching computational chemistry is now possible, enabling students to perform reasonable modeling on their own computing devices, in the bring your own device 1 (BYOD) scheme. In addition to the programs’ use for various applications, open access to the programs’ source code also enables comprehensive teaching strategies, as actual algorithms’ implementations can be used in teaching. -

User Guide of Computational Facilities
Physical Chemistry Unit Departament de Quimica User Guide of Computational Facilities Beta Version System Manager: Marc Noguera i Julian January 20, 2009 Contents 1 Introduction 1 2 Computer resources 2 3 Some Linux tools 2 4 The Cluster 2 4.1 Frontend servers . 2 4.2 Filesystems . 3 4.3 Computational nodes . 3 4.4 User’s environment . 4 4.5 Disk space quota . 4 4.6 Data Backup . 4 4.7 Generation of SSH keys for automatic SSH login . 4 4.8 Submitting jobs to the cluster . 5 4.8.1 Typical queue system commands . 6 4.8.2 HomeMade scripts . 6 4.8.3 Create your own submit script . 6 4.8.4 Submitting Parallel Calculations . 7 5 Available Software 9 5.1 Chemisty software . 9 5.2 Development Software . 10 5.3 How to use the software . 11 5.4 Non-default environments . 11 6 Your Linux Workstation 11 6.1 Disk space on your workstation . 12 6.2 Workstation Data Backup . 12 6.3 Running virtual Windows XP . 13 6.3.1 The Windows XP Environment . 13 6.4 Submitting calculations to your dekstop . 13 6.5 Network dependent structure . 14 7 Your WindowsXP Workstation 14 7.1 Workstation Data Backup . 14 8 Frequently asked questions 14 1 Introduction This User Guide is aimed to the standard user of the computer facilities in the ”Unitat de Quimica Fisica” in the Universitat Autnoma de Barcelona. It is not assumed that you have a knowledge of UNIX/Linux. However, you are 1 encouraged to take a close look to the linux guides that will be pointed out. -

D6.1 Report on the Deployment of the Max Demonstrators and Feedback to WP1-5
Ref. Ares(2020)2820381 - 31/05/2020 HORIZON2020 European Centre of Excellence Deliverable D6.1 Report on the deployment of the MaX Demonstrators and feedback to WP1-5 D6.1 Report on the deployment of the MaX Demonstrators and feedback to WP1-5 Pablo Ordejón, Uliana Alekseeva, Stefano Baroni, Riccardo Bertossa, Miki Bonacci, Pietro Bonfà, Claudia Cardoso, Carlo Cavazzoni, Vladimir Dikan, Stefano de Gironcoli, Andrea Ferretti, Alberto García, Luigi Genovese, Federico Grasselli, Anton Kozhevnikov, Deborah Prezzi, Davide Sangalli, Joost VandeVondele, Daniele Varsano, Daniel Wortmann Due date of deliverable: 31/05/2020 Actual submission date: 31/05/2020 Final version: 31/05/2020 Lead beneficiary: ICN2 (participant number 3) Dissemination level: PU - Public www.max-centre.eu 1 HORIZON2020 European Centre of Excellence Deliverable D6.1 Report on the deployment of the MaX Demonstrators and feedback to WP1-5 Document information Project acronym: MaX Project full title: Materials Design at the Exascale Research Action Project type: European Centre of Excellence in materials modelling, simulations and design EC Grant agreement no.: 824143 Project starting / end date: 01/12/2018 (month 1) / 30/11/2021 (month 36) Website: www.max-centre.eu Deliverable No.: D6.1 Authors: P. Ordejón, U. Alekseeva, S. Baroni, R. Bertossa, M. Bonacci, P. Bonfà, C. Cardoso, C. Cavazzoni, V. Dikan, S. de Gironcoli, A. Ferretti, A. García, L. Genovese, F. Grasselli, A. Kozhevnikov, D. Prezzi, D. Sangalli, J. VandeVondele, D. Varsano, D. Wortmann To be cited as: Ordejón, et al., (2020): Report on the deployment of the MaX Demonstrators and feedback to WP1-5. Deliverable D6.1 of the H2020 project MaX (final version as of 31/05/2020). -

Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development
processes Review Molecular Dynamics Simulations in Drug Discovery and Pharmaceutical Development Outi M. H. Salo-Ahen 1,2,* , Ida Alanko 1,2, Rajendra Bhadane 1,2 , Alexandre M. J. J. Bonvin 3,* , Rodrigo Vargas Honorato 3, Shakhawath Hossain 4 , André H. Juffer 5 , Aleksei Kabedev 4, Maija Lahtela-Kakkonen 6, Anders Støttrup Larsen 7, Eveline Lescrinier 8 , Parthiban Marimuthu 1,2 , Muhammad Usman Mirza 8 , Ghulam Mustafa 9, Ariane Nunes-Alves 10,11,* , Tatu Pantsar 6,12, Atefeh Saadabadi 1,2 , Kalaimathy Singaravelu 13 and Michiel Vanmeert 8 1 Pharmaceutical Sciences Laboratory (Pharmacy), Åbo Akademi University, Tykistökatu 6 A, Biocity, FI-20520 Turku, Finland; ida.alanko@abo.fi (I.A.); rajendra.bhadane@abo.fi (R.B.); parthiban.marimuthu@abo.fi (P.M.); atefeh.saadabadi@abo.fi (A.S.) 2 Structural Bioinformatics Laboratory (Biochemistry), Åbo Akademi University, Tykistökatu 6 A, Biocity, FI-20520 Turku, Finland 3 Faculty of Science-Chemistry, Bijvoet Center for Biomolecular Research, Utrecht University, 3584 CH Utrecht, The Netherlands; [email protected] 4 Swedish Drug Delivery Forum (SDDF), Department of Pharmacy, Uppsala Biomedical Center, Uppsala University, 751 23 Uppsala, Sweden; [email protected] (S.H.); [email protected] (A.K.) 5 Biocenter Oulu & Faculty of Biochemistry and Molecular Medicine, University of Oulu, Aapistie 7 A, FI-90014 Oulu, Finland; andre.juffer@oulu.fi 6 School of Pharmacy, University of Eastern Finland, FI-70210 Kuopio, Finland; maija.lahtela-kakkonen@uef.fi (M.L.-K.); tatu.pantsar@uef.fi -
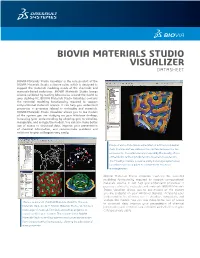
Biovia Materials Studio Visualizer Datasheet
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

Download Author Version (PDF)
PCCP Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/pccp Page 1 of 11 PhysicalPlease Chemistry do not adjust Chemical margins Physics PCCP PAPER Effect of nanosize on surface properties of NiO nanoparticles for adsorption of Quinolin-65 ab a a Received 00th January 20xx, Nedal N. Marei, Nashaat N. Nassar* and Gerardo Vitale Accepted 00th January 20xx Using Quinolin-65 (Q-65) as a model-adsorbing compound for polar heavy hydrocarbons, the nanosize effect of NiO Manuscript DOI: 10.1039/x0xx00000x nanoparticles on adsorption of Q-65 was investigated. Different-sized NiO nanoparticles with sizes between 5 and 80 nm were prepared by controlled thermal dehydroxylation of Ni(OH)2. -

Parameterizing a Novel Residue
University of Illinois at Urbana-Champaign Luthey-Schulten Group, Department of Chemistry Theoretical and Computational Biophysics Group Computational Biophysics Workshop Parameterizing a Novel Residue Rommie Amaro Brijeet Dhaliwal Zaida Luthey-Schulten Current Editors: Christopher Mayne Po-Chao Wen February 2012 CONTENTS 2 Contents 1 Biological Background and Chemical Mechanism 4 2 HisH System Setup 7 3 Testing out your new residue 9 4 The CHARMM Force Field 12 5 Developing Topology and Parameter Files 13 5.1 An Introduction to a CHARMM Topology File . 13 5.2 An Introduction to a CHARMM Parameter File . 16 5.3 Assigning Initial Values for Unknown Parameters . 18 5.4 A Closer Look at Dihedral Parameters . 18 6 Parameter generation using SPARTAN (Optional) 20 7 Minimization with new parameters 32 CONTENTS 3 Introduction Molecular dynamics (MD) simulations are a powerful scientific tool used to study a wide variety of systems in atomic detail. From a standard protein simulation, to the use of steered molecular dynamics (SMD), to modelling DNA-protein interactions, there are many useful applications. With the advent of massively parallel simulation programs such as NAMD2, the limits of computational anal- ysis are being pushed even further. Inevitably there comes a time in any molecular modelling scientist’s career when the need to simulate an entirely new molecule or ligand arises. The tech- nique of determining new force field parameters to describe these novel system components therefore becomes an invaluable skill. Determining the correct sys- tem parameters to use in conjunction with the chosen force field is only one important aspect of the process. -

Supporting Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information A Single Crystalline Porphyrinic Titanium Metal−Organic Framework Shuai Yuana†, Tian-Fu Liua†, Dawei Fenga, Jian Tiana, Kecheng Wanga, Junsheng Qina, a a a a b Qiang Zhang , Ying-Pin Chen , Mathieu Bosch , Lanfang Zou , Simon J. Teat, Scott J. c a Dalgarno and Hong-Cai Zhou * a Department of Chemistry, Texas A&M University, College Station, Texas 77842-3012, USA b Advanced Light Source, Lawrence Berkeley National Laboratory Berkeley, CA 94720, USA c Institute of Chemical Sciences, Heriot-Watt University Riccarton, Edinburgh EH14 4AS, U.K. † Equal contribution to this work *To whom correspondence should be addressed. Email: [email protected] Tel: +1 (979) 845-4034; Fax: +1 (979) 845-1595 S1 Contents S1. Ligand Synthesis..............................................................................................................3 S2. Syntheses of PCN-22.......................................................................................................5 S3. X-ray Crystallography .....................................................................................................6 S4. Topological Analyses ......................................................................................................9 S5. N2 Sorption Isotherm .....................................................................................................10 S6. Simulation of the Accessible Surface Area ...................................................................12 -

3D-Printing Models for Chemistry
3D-Printing Models for Chemistry: A Step-by-Step Open-Source Guide for Hobbyists, Corporate ProfessionAls, and Educators and Student in K-12 and Higher Education Poster Elisabeth Grace Billman-Benveniste+, Jacob Franz++, Loredana Valenzano-Slough+* +Department of Chemistry, Michigan Technological University, ++Department of Mechanical Engineering, Michigan Technological University *Corresponding Author References 1. “LulzBot TAZ 5.” LulzBot, 14 Aug. 2018, www.lulzbot.com/store/printers/lulzbot-taz-5 2. Gaussian 16, Revision B.01, Frisch, M. J.; Trucks, G. W.; Schlegel, H. B.; Scuseria, G. E.; Robb, M. A.; Cheeseman, J. R.; Scalmani, G.; Barone, V.; Petersson, G. A.; Nakatsuji, H.; Li, X.; Caricato, M.; Marenich, A. V.; Bloino, J.; Janesko, B. G.; Gomperts, R.; Mennucci, B.; Hratchian, H. P.; Ortiz, J. V.; Izmaylov, A. F.; Sonnenberg, J. L.; Williams-Young, D.; Ding, F.; Lipparini, F.; Egidi, F.; Goings, J.; Peng, B.; Petrone, A.; Henderson, T.; Ranasinghe, D.; ZakrzeWski, V. G.; Gao, J.; Rega, N.; Zheng, G.; Liang, W.; Hada, M.; Ehara, M.; Toyota, K.; Fukuda, R.; HasegaWa, J.; Ishida, M.; NakaJima, T.; Honda, Y.; Kitao, O.; Nakai, H.; Vreven, T.; Throssell, K.; Montgomery, J. A., Jr.; Peralta, J. E.; Ogliaro, F.; Bearpark, M. J.; Heyd, J. J.; Brothers, E. N.; Kudin, K. N.; Staroverov, V. N.; Keith, T. A.; Kobayashi, R.; Normand, J.; Raghavachari, K.; Rendell, A. P.; Burant, J. C.; Iyengar, S. S.; Tomasi, J.; Cossi, M.; Millam, J. M.; Klene, M.; Adamo, C.; Cammi, R.; Ochterski, J. W.; Martin, R. L.; Morokuma, K.; Farkas, O.; Foresman, J. B.; Fox, D. J. Gaussian, Inc., Wallingford CT, 2016. 3. -

Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins
Article Reactive Molecular Dynamics Study of the Thermal Decomposition of Phenolic Resins Marcus Purse 1, Grace Edmund 1, Stephen Hall 1, Brendan Howlin 1,* , Ian Hamerton 2 and Stephen Till 3 1 Department of Chemistry, Faculty of Engineering and Physical Sciences, University of Surrey, Guilford, Surrey GU2 7XH, UK; [email protected] (M.P.); [email protected] (G.E.); [email protected] (S.H.) 2 Bristol Composites Institute (ACCIS), Department of Aerospace Engineering, School of Civil, Aerospace, and Mechanical Engineering, University of Bristol, Bristol BS8 1TR, UK; [email protected] 3 Defence Science and Technology Laboratory, Porton Down, Salisbury SP4 0JQ, UK; [email protected] * Correspondence: [email protected]; Tel.: +44-1483-686-248 Received: 6 March 2019; Accepted: 23 March 2019; Published: 28 March 2019 Abstract: The thermal decomposition of polyphenolic resins was studied by reactive molecular dynamics (RMD) simulation at elevated temperatures. Atomistic models of the polyphenolic resins to be used in the RMD were constructed using an automatic method which calls routines from the software package Materials Studio. In order to validate the models, simulated densities and heat capacities were compared with experimental values. The most suitable combination of force field and thermostat for this system was the Forcite force field with the Nosé–Hoover thermostat, which gave values of heat capacity closest to those of the experimental values. Simulated densities approached a final density of 1.05–1.08 g/cm3 which compared favorably with the experimental values of 1.16–1.21 g/cm3 for phenol-formaldehyde resins.