Hereditary Fructose Intolerance Caused by a Nonsense Mutation Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Targeted Therapies for Metabolic Myopathies Related to Glycogen Storage and Lipid Metabolism: a Systematic Review and Steps Towards a ‘Treatabolome’
Journal of Neuromuscular Diseases 8 (2021) 401–417 401 DOI 10.3233/JND-200621 IOS Press Systematic Review Targeted Therapies for Metabolic Myopathies Related to Glycogen Storage and Lipid Metabolism: a Systematic Review and Steps Towards a ‘Treatabolome’ A. Mantaa,b, S. Spendiffb, H. Lochmuller¨ b,c,d,e,f and R. Thompsonb,∗ aFaculty of Medicine, University of Ottawa, Ottawa, ON, Canada bChildren’s Hospital of Eastern Ontario Research Institute, Ottawa, ON, Canada cDepartment of Neuropediatrics and Muscle Disorders, Medical Center – University of Freiburg, Faculty of Medicine, Freiburg, Germany dCentro Nacional de An´alisis Gen´omico (CNAG-CRG), Center for Genomic Regulation, Barcelona Institute of Science and Technology (BIST), Barcelona, Catalonia, Spain eDivision of Neurology, Department of Medicine, The Ottawa Hospital, University of Ottawa, Ottawa, Canada f Brain and Mind Research Institute, University of Ottawa, Ottawa, Canada Abstract. Background: Metabolic myopathies are a heterogenous group of muscle diseases typically characterized by exercise intoler- ance, myalgia and progressive muscle weakness. Effective treatments for some of these diseases are available, but while our understanding of the pathogenesis of metabolic myopathies related to glycogen storage, lipid metabolism and -oxidation is well established, evidence linking treatments with the precise causative genetic defect is lacking. Objective: The objective of this study was to collate all published evidence on pharmacological therapies for the aforemen- tioned metabolic myopathies and link this to the genetic mutation in a format amenable to databasing for further computational use in line with the principles of the “treatabolome” project. Methods: A systematic literature review was conducted to retrieve all levels of evidence examining the therapeutic efficacy of pharmacological treatments on metabolic myopathies related to glycogen storage and lipid metabolism. -

Datasheet: VPA00226
Datasheet: VPA00226 Description: RABBIT ANTI ALDOA Specificity: ALDOA Format: Purified Product Type: PrecisionAb™ Polyclonal Isotype: Polyclonal IgG Quantity: 100 µl Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Species Cross Reacts with: Mouse, Rat Reactivity N.B. Antibody reactivity and working conditions may vary between species. Product Form Purified IgG - liquid Preparation Rabbit Ig fraction prepared by ammonium sulphate precipitation Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers Immunogen KLH conjugated synthetic peptide between 66-95 amino acids from the N-terminal region of human ALDOA External Database UniProt: Links P04075 Related reagents Entrez Gene: 226 ALDOA Related reagents Page 1 of 2 Synonyms ALDA Specificity Rabbit anti Human ALDOA antibody recognizes fructose-bisphosphate aldolase A, also known as epididymis secretory sperm binding protein Li 87p, fructose-1,6-bisphosphate triosephosphate-lyase, lung cancer antigen NY-LU-1 and muscle-type aldolase. Encoded by the ALDOA gene, fructose-bisphosphate aldolase A is a glycolytic enzyme that catalyzes the reversible conversion of fructose-1,6-bisphosphate to glyceraldehyde 3-phosphate and dihydroxyacetone phosphate. -

Maladies Convention Mal
maladies métaboliques monogéniques héréditaires rares 1 avenant à la convention avec les centres de référence – annexe OMIM # FULL NAME A. disorders of amino acid metabolism 1 261600 classical phenylketonuria and hyperphenylala- ninemia 2 261640 phenylketonuria due to PTPS deficiency 3 261630 phenylketonuria due to DHPR deficiency 4 264070 phenylketonuria due to PCD deficiency 5 128230 DOPA-responsive dystonia (TH, SPR, GCH1) 6 248600 leucinose, maple syrup urine disease (MSUD) 7 276700 tyrosinemia type 1 8 276600 tyrosinemia type 2 9 276710 tyrosinemia type 3 10 203500 alkaptonuria 11 236200 homocystinuria, B6 responsive and non responsive 12 236250 homocystinuria due to MTHFR deficiency 13 236270-250940 homocystinuria-megaloblastic anemia Cbl E & G type 14 250850 methionine S-adenosyltransferase deficiency 15 606664 glycine N-methyltransferase deficiency 16 180960 S-adenosylhomocystine hydrolase deficiency 17 237300 hyperammonemia due to CPS deficiency 18 311250 hyperammonemia due to OTC deficiency 19 215700 citrullinemia type I 20 605814-603471 citrullinemia type II 21 207900 argininosuccinic aciduria (ASL deficiency) 22 207800 argininemia (arginase deficiency) 23 237310 hyperammonemia due to NAGS deficiency 24 238970 hyperornithinemia, hyperammonemia, homocitrulli- nuria (HHH) 25 222700 lysinuric protein intolerance 26 258870 gyrate atrophy, B6 responsive or non responsive 27 238700 hyperlysinemia (alpha-aminoadipic semialdehyde synthase deficiency) 28 238300-330-310 non ketotic hyperglycinaemia 29 234500 hartnup disorder 30 601815-172480 -

Table S1. Disease Classification and Disease-Reaction Association
Table S1. Disease classification and disease-reaction association Disorder class Associated reactions cross Disease Ref[Goh check et al. -

The Stochastic Human Red Blood Cell Model and Its Applications
RESEARCH The Stochastic Human Red Blood Cell Model and Its Applications Archita Biswas1, Pawan K Dhar1, Girja Kaushal2, Usha Kumari3, Kalaiarasan Ponnusamy* 1Synthetic Biology group, School of Biotechnology, Jawaharlal Nehru University, New Delhi, India 2Center of Innovative and Applied Bioprocessing, Mohali, Punjab, India 3Faculty of Medicine, AIMST University, Malaysia. *Corresponding author’s email: [email protected] ABSTRACT Red blood cells (RBCs) form the bulk of the blood volume for delivering oxygen and nutrients to various cells in the body. Several studies have focused on the RBCs to understand its metabolic activities in normal and abnormal conditions. However, an integrated view of the red blood cell, as a system of molecular fluctuations, is still lacking. The RBC metabolic wiring comprises four key pathways, i.e. Glycolysis, Pentose Phosphate, Purine salvage pathway and Glutathione pathway. Here we present an integrated metabolic pathway model of the human red blood cell by including first three pathways due to their significant presence and interaction. Furthermore, the stochastic model of RBCs metabolic pathways has been extended to include explaining abnormal conditions in the form of Pyruvate Kinase and Aldolase A deficiencies. By studying metabolite changes in-silico, four biomarkers were found to represent two genetic mutations, i.e., 2-phosphoglycerate & phosphoenol pyruvate changes for Pyruvate kinase deficiency and fructose-1-6-bisphosphate & xylulose-5-phosphate for Aldolase A deficiency. The virtual RBC model has the potential for more clinical applications and can be scaled up to include additional regulatory and metabolic content. KEYWORDS: Virtual Cell, stochastic model, RBC, ns, Pyruvate kinase, Aldolase A deficiency. Citation: Biswas A et al (2018) The Stochastic human red blood cell model and its applications. -

6 the Glycogen Storage Diseases and Related Disorders
6 The Glycogen Storage Diseases and Related Disorders G. Peter A. Smit, Jan Peter Rake, Hasan O. Akman, Salvatore DiMauro 6.1 The Liver Glycogenoses – 103 6.1.1 Glycogen Storage Disease Type I (Glucose-6-Phosphatase or Translocase Deficiency) – 103 6.1.2 Glycogen Storage Disease Type III (Debranching Enzyme Deficiency) – 108 6.1.3 Glycogen Storage Disease Type IV (Branching Enzyme Deficiency) – 109 6.1.4 Glycogen Storage Disease Type VI (Glycogen Phosphorylase Deficiency) – 111 6.1.5 Glycogen Storage Disease Type IX (Phosphorylase Kinase Deficiency) – 111 6.1.6 Glycogen Storage Disease Type 0 (Glycogen Synthase Deficiency) – 112 6.2 The Muscle Glycogenoses – 112 6.2.1 Glycogen Storage Disease Type V (Myophosphorylase Deficiency) – 113 6.2.2 Glycogen Storage Disease Type VII (Phosphofructokinase Deficiency) – 113 6.2.3 Phosphoglycerate Kinase Deficiency – 114 6.2.4 Glycogen Storage Disease Type X (Phosphoglycerate Mutase Deficiency) – 114 6.2.5 Glycogen Storage Disease Type XII (Aldolase A Deficiency) – 114 6.2.6 Glycogen Storage Disease Type XIII (E-Enolase Deficiency) – 115 6.2.7 Glycogen Storage Disease Type XI (Lactate Dehydrogenase Deficiency) – 115 6.2.8 Muscle Glycogen Storage Disease Type 0 (Glycogen Synthase Deficiency) – 115 6.3 The Generalized Glycogenoses and Related Disorders – 115 6.3.1 Glycogen Storage Disease Type II (Acid Maltase Deficiency) – 115 6.3.2 Danon Disease – 116 6.3.3 Lafora Disease – 116 References – 116 102 Chapter 6 · The Glycogen Storage Diseases and Related Disorders Glycogen Metabolism Glycogen is a macromolecule composed of glucose viding glucose and glycolytic intermediates (. Fig. 6.1). units. -

Aldolase Activity Colorimetric Assay Kit (Catalog # K665-100; 100 Assays; Store at -20°C) 4/14
FOR RESEARCH USE ONLY! Aldolase Activity Colorimetric Assay Kit (Catalog # K665-100; 100 assays; Store at -20°C) 4/14 I. Introduction: Aldolase (Fructose-Bisphosphate Aldolase: EC 4.1.2.13) is an important enzyme for both glycolysis and gluconeogenesis. It catalyzes the reversible reaction of fructose-1,6-bisphosphate to glyceraldehyde-3-phosphate & dihydroxyacetone. There are 2 classes of Aldolase - class l: found in animal and plant tissues and class II: found in prokaryotes and lower eukaryotes. Class I Aldolase has 3 isozymes- Type A: found in muscle and red blood cells, Type B: found in liver and kidney and Type C: found in brain. Aldolase A deficiency leads to myopathy & hemolytic anemia. Muscle disease and liver injury can also cause increased serum aldolase. Accurate detection of aldolase activity is valuable for diagnostic and mechanistic studies. In BioVision’s Aldolase Activity Assay, aldolase converts fructose-1,6- bisphosphate to glyceraldehyde-3-phosphate and dihydroxyacetone, and through a series of reactions, reduces a nearly colorless probe to a colored product with absorbance at 450 nm. This assay kit is simple, sensitive, and high-throughput adaptable. Detection limit: less than 0.1 mU of aldolase activity in a variety of samples. Aldolase Fructose-1,6-Bisphosphate Glyceraldehyde -3-Phosphate + Dihydroxyacetone Phosphate Color (OD 450 nm) II. Applications: Measurement of Aldolase activity in various tissues/cells Analysis of glycolysis and gluconeogenesis pathways III. Sample Type: Animal tissues: muscle, liver, heart, kidney, etc. Cell culture: adherent or suspension cells Human serum and plasma IV. Kit Contents: Components K665-100 Cap Code Part Number Aldolase Assay Buffer 25 ml WM K665-100-1 Aldolase Substrate (Lyophilized) 1 vial Blue K665-100-2 Aldolase Enzyme Mix (Lyophilized) 1 vial Green K665-100-3 Aldolase Developer (Lyophilized) 1 vial Red K665-100-4 NADH Standard (Lyophilized) 1 vial Yellow K665-100-5 Aldolase Positive Control (Lyophilized) 1 vial Orange K665-100-6 V. -

15 Disorders of Carbohydrate and Glycogen Metabolism Jan Peter Rake, Gepke Visser, G
15 Disorders of Carbohydrate and Glycogen Metabolism Jan Peter Rake, Gepke Visser, G. Peter A. Smit 15.1 Introduction The disorders described in this chapter have symptoms varying from mild to severe and life-threatening. The symptoms comprise failure to thrive, hep- atomegaly, jaundice and liver failure, hypoglycemia, metabolic acidosis, and (cardio-) myopathy, including muscle pain and exercise intolerance. Four groups of disorders can be distinguished: A. Disorders of galactose metabolism comprise galactokinase deficiency, gal- actose-1-P-uridyl transferase deficiency (classical galactosemia), and UDP- galactose-4-epimerase deficiency. The primary source of dietary galactose is lactose, the sugar in milk. It is present in human and cow’s milk and in most infant formulae. Individuals with one of these enzyme defects are unable to transform galactose into glucose and they accumulate metabolites of galactose after ingesting lactose and/or galactose. Galactitol accumulation accounts for cataract formation. Galactose-1-phosphate is considered to be responsible for the other clinical manifestations, especially liver and kidney failure. Cataracts are the only manifestations of galactose kinase deficiency. The clinical manifestations of classic galactosemia are vomiting, failure to thrive, liver failure with jaundice, kidney failure, cataract, and sepsis, occurring when galactose is introduced in the diet. The severe form of UDP galacose-4-epimerase deficiency resembles classic galactosemia. The main goal of treatment for galactokinase deficiency (more liberal) and classic galactosemia (strict) is the elimination of galactose from the diet. In severe forms of UDP galactose-4-epimerase deficiency, a narrow balance in dietary galactose requirements for biosynthesis (galactosylated compounds) and excess causing accumulation of galactose-1-phosphate should be aimed for. -
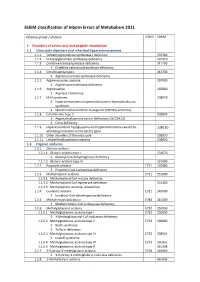
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

ORPHANET 3 (Phase 3)
Programme of community action on rare diseases Contract 2002/CVG4 ORPHANET : Intermediate scientific report May 2003 Summary The project was to extend the content of the already existing ORPHANET database to build up a truly European database. The first year (Dec 00-November 01) was the feasibility study year and a pilot study with four countries. The second year (Dec 01- Nov 02) was the year of the move from a French encyclopaedia to a European one, and the year of the collection of data on services in 7 countries. The third year (Dec 1,2002 – Nov 03) is the year of the data collection up to completeness in 7 of the participating countries and the year of identification of sources and satrt of the data collection in the new country: Portugal.. For the encyclopaedia, a board of 83 editors has been established progressively, specialty by specialty and authors of texts nominated. For the 3,500 diseases, there are on-line: 990 summaries in French, 833 summaries in English, 445 review articles in French or in English. The data about services are partially collected in all participating countries and already released for Italy, Belgium, Switzerland, Germany and Spain. The amount of data released is: 594 patient support groups, 945 laboratories providing diagnostic tests, 1392 research projects and 945 expert clinics. The Italian, German and Spanish versions of the website are now active. The usefulness of the database is assessed through the number of connections. In April 2003, we have had during the month visits from 101,400 different visits from 113 different countries. -

A Thermolabile Aldolase a Mutant Causes Fever-Induced Recurrent Rhabdomyolysis Without Hemolytic Anemia
A Thermolabile Aldolase A Mutant Causes Fever-Induced Recurrent Rhabdomyolysis without Hemolytic Anemia Asmaa Mamoune1, Michel Bahuau2, Yamina Hamel1, Vale´rie Serre3, Michele Pelosi1, Florence Habarou4, Marie-Ange Nguyen Morel5, Bertrand Boisson6, Sabrina Vergnaud7, Mai Thao Viou8, Luc Nonnenmacher8, Monique Piraud9, Patrick Nusbaum10, Joseph Vamecq11, Norma Romero7, Chris Ottolenghi8, Jean-Laurent Casanova5,12, Pascale de Lonlay1* 1 INSERM U781, Institut Imagine des Maladies Ge´ne´tiques, Universite´ Paris Descartes et Centre de Re´fe´rence des Maladies He´re´ditaires du Me´tabolisme, Hoˆpital Necker, AP-HP, Paris, France, 2 De´partement de Ge´ne´tique, Hoˆpitaux Universitaires Henri-Mondor, Cre´teil, AP-HP, France, 3 "Mitochondria, Metals and Oxidative Stress" group, Jacques Monod Institute, UMR7592 CNRS, Paris Diderot University, Paris, France, 4 Metabolic biochemistry and INSERM U1124, University Paris Descartes, Hospital Necker Enfants Malades, Paris, France, 5 Clinique Universitaire de Pe´diatrie, Hoˆpital couple enfant, CHU de Grenoble, France, 6 St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, New York, United States of America; Unite´ Institut National de la Sante´ et de la Recherche Me´dicale U980, Laboratory of Human Genetics of Infectious Diseases, Imagine Institute; and Pediatric Hematology-Immunology Unit, Necker Hospital for Sick Children, University Paris Descartes, Paris, France, 7 De´partement de Biochimie, Toxicologie et Pharmacologie, CHU de Grenoble, -

Glycogen Storage Diseases the Patient-Parent Handbook
Glycogen Storage Diseases The Patient-Parent Handbook AGSD’s “Glycogen Storage Diseases: A Patient-Parent Handbook” TABLE OF CONTENTS Chapter 1 The Biochemistry of Glycogen Storage Disease ………………………………………………3 Chapter 2 Important Terms …………………………………………………………………………….…………….7 Chapter 3 Glycogen Storage Diseases ……………………………………………………………………………11 Chapter 4 Type I Glycogen Storage Disease ………………………………………………………………...…13 Chapter 5 Type II Glycogen Storage Disease (abbreviated GSD II) …………………………………..21 Chapter 6 Type III Glycogen Storage Disease (abbreviated GSD III) ………………………………..25 Chapter 7 Type IV Glycogen Storage Disease (abbreviated GSD IV) ………………………………..28 Chapter 8 McArdle Disease …………………………………………………………………………………………..30 Chapter 9 Type VI Glycogen Storage Disease (abbreviated GSD VI) and Type IX Glycogen Storage Disease (abbreviated GSD IX …………………………………………………………………………….33 Chapter 10 Type VII Glycogen Storage Disease (abbreviated GSD VII) ……………………………36 Chapter 11 Type 0 Glycogen Storage Disease (abbreviated GSD 0) …………………………………38 Chapter 12 Newer Glycogen Storage Diseases ……………………………………………………………….40 Chapter 13 Parent, Family and Patient Involvement ………………………………………………………42 Chapter 14 Questions and Answers ………………………………………………………………………………44 Chapter 15 Where to Get Information …………………………………………………………………………...48 Chapter 16 Glossary of Terms Related to Glycogen Storage Disease ……………………………….49 2 AGSD’s “Glycogen Storage Diseases: A Patient-Parent Handbook” Chapter 1 The Biochemistry of Glycogen Storage Disease The underlying problem in all of the glycogen storage diseases is the use and storage of glycogen. Glycogen is a complex material composed of glucose molecules linked together. HOW THE BODY STORES GLUCOSE AS GLYCOGEN Glucose is a basic sugar (see Figure 1). It is an important source of energy for the body. and is the main transport form of energy in the blood stream. The body usually keeps the level of glucose in the blood within a narrow range of concentrations: 60-100 units (mg per deciliters).