Epstein–Barr Virus Nuclear Antigen 3C Regulated Genes in Lymphoblastoid Cell Lines
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Specific Functions of the Wnt Signaling System in Gene Regulatory Networks
Specific functions of the Wnt signaling system in PNAS PLUS gene regulatory networks throughout the early sea urchin embryo Miao Cui, Natnaree Siriwon1, Enhu Li2, Eric H. Davidson3, and Isabelle S. Peter3 Division of Biology and Biological Engineering, California Institute of Technology, Pasadena, CA 91125 Contributed by Eric H. Davidson, October 9, 2014 (sent for review September 12, 2014; reviewed by Robert D. Burke and Randall T. Moon) Wnt signaling affects cell-fate specification processes throughout Fig. 1A. Cells located at the vegetal pole will become skeleto- embryonic development. Here we take advantage of the well-studied genic mesodermal cells. These cells are surrounded by the veg2 gene regulatory networks (GRNs) that control pregastrular sea urchin cell lineage. This lineage consists of veg2 mesodermal cells, lo- embryogenesis to reveal the gene regulatory functions of the entire cated adjacent to skeletogenic cells and giving rise to all other Wnt-signaling system. Five wnt genes, three frizzled genes, two se- mesodermal cell fates such as esophageal muscle cells, blasto- creted frizzled-related protein 1 genes, and two Dickkopf genes are coelar cells, and pigment cells, and of veg2 endoderm cells, expressed in dynamic spatial patterns in the pregastrular embryo of which will form the foregut and parts of the midgut. At a further Strongylocentrotus purpuratus. We present a comprehensive analysis distance from the vegetal pole, but still within the vegetal half of of these genes in each embryonic domain. Total functions of the the embryo, is the veg1 lineage, consisting of veg1 endoderm, Wnt-signaling system in regulatory gene expression throughout the located adjacent to veg2 endoderm and giving rise to the other embryo were studied by use of the Porcupine inhibitor C59, which parts of the midgut and the hindgut, and of veg1 ectoderm, the interferes with zygotic Wnt ligand secretion. -

Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 T + is online at: average * The Journal of Immunology , 34 of which you can access for free at: 2016; 197:1477-1488; Prepublished online 1 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600589 http://www.jimmunol.org/content/197/4/1477 Molecular Profile of Tumor-Specific CD8 Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. Waugh, Sonia M. Leach, Brandon L. Moore, Tullia C. Bruno, Jonathan D. Buhrman and Jill E. Slansky J Immunol cites 95 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2016/07/01/jimmunol.160058 9.DCSupplemental This article http://www.jimmunol.org/content/197/4/1477.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Molecular Profile of Tumor-Specific CD8+ T Cell Hypofunction in a Transplantable Murine Cancer Model Katherine A. -

Rb-Mediated Neuronal Differentiation Through Cell-Cycle–Independent Regulation of E2f3a
PLoS BIOLOGY Rb-Mediated Neuronal Differentiation through Cell-Cycle–Independent Regulation of E2f3a Danian Chen1,2,3, Rene Opavsky4,5,6, Marek Pacal1,2,3, Naoyuki Tanimoto7, Pamela Wenzel4,5,6, Mathias W. Seeliger7, Gustavo Leone4,5,6, Rod Bremner1,2,3* 1 Genetics and Development Division, Toronto Western Research Institute, University Health Network, University of Toronto, Ontario, Canada, 2 Department of Ophthalmology and Visual Science, University of Toronto, Ontario, Canada, 3 Department of Laboratory Medicine and Pathobiology, University of Toronto, Ontario, Canada, 4 Human Cancer Genetics Program, Department of Molecular Virology, Immunology and Medical Genetics, Ohio State University, Columbus, Ohio, United States of America, 5 Department of Molecular Genetics, Ohio State University, Columbus, Ohio, United States of America, 6 Comprehensive Cancer Center, Ohio State University, Columbus, Ohio, United States of America, 7 Ocular Neurodegeneration Research Group, Centre for Ophthalmology, Institute for Ophthalmic Research, University of Tuebingen, Germany It has long been known that loss of the retinoblastoma protein (Rb) perturbs neural differentiation, but the underlying mechanism has never been solved. Rb absence impairs cell cycle exit and triggers death of some neurons, so differentiation defects may well be indirect. Indeed, we show that abnormalities in both differentiation and light- evoked electrophysiological responses in Rb-deficient retinal cells are rescued when ectopic division and apoptosis are blocked specifically by deleting E2f transcription factor (E2f) 1. However, comprehensive cell-type analysis of the rescued double-null retina exposed cell-cycle–independent differentiation defects specifically in starburst amacrine cells (SACs), cholinergic interneurons critical in direction selectivity and developmentally important rhythmic bursts. Typically, Rb is thought to block division by repressing E2fs, but to promote differentiation by potentiating tissue- specific factors. -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -

Reversal of Glucocorticoid Resistance in Paediatric Acute Lymphoblastic Leukaemia Is Dependent on Restoring BIM Expression
www.nature.com/bjc ARTICLE Translational Therapeutics Reversal of glucocorticoid resistance in paediatric acute lymphoblastic leukaemia is dependent on restoring BIM expression Cara E. Toscan1, Duohui Jing1, Chelsea Mayoh1 and Richard B. Lock1 BACKGROUND: Acute lymphoblastic leukaemia (ALL) is the most common paediatric malignancy. Glucocorticoids form a critical component of chemotherapy regimens and resistance to glucocorticoid therapy is predictive of poor outcome. We have previously shown that glucocorticoid resistance is associated with upregulation of the oncogene C-MYC and failure to induce the proapoptotic gene BIM. METHODS: A high-throughput screening (HTS) campaign was carried out to identify glucocorticoid sensitisers against an ALL xenograft derived from a glucocorticoid-resistant paediatric patient. Gene expression analysis was carried out using Illumina microarrays. Efficacy, messenger RNA and protein analysis were carried out by Resazurin assay, reverse transcription-PCR and immunoblotting, respectively. RESULTS: A novel glucocorticoid sensitiser, 2-((4,5-dihydro-1H-imidazol-2-yl)thio)-N-isopropyl-N-phenylacetamide (GCS-3), was identified from the HTS campaign. The sensitising effect was specific to glucocorticoids and synergy was observed in a range of dexamethasone-resistant and dexamethasone-sensitive xenografts representative of B-ALL, T-ALL and Philadelphia chromosome- positive ALL. GCS-3 in combination with dexamethasone downregulated C-MYC and significantly upregulated BIM expression in a glucocorticoid-resistant ALL xenograft. The GCS-3/dexamethasone combination significantly increased binding of the glucocorticoid receptor to a novel BIM enhancer, which is associated with glucocorticoid sensitivity. CONCLUSIONS: This study describes the potential of the novel glucocorticoid sensitiser, GCS-3, as a biological tool to interrogate glucocorticoid action and resistance. -

Tgfβ-Regulated Gene Expression by Smads and Sp1/KLF-Like Transcription Factors in Cancer VOLKER ELLENRIEDER
ANTICANCER RESEARCH 28 : 1531-1540 (2008) Review TGFβ-regulated Gene Expression by Smads and Sp1/KLF-like Transcription Factors in Cancer VOLKER ELLENRIEDER Signal Transduction Laboratory, Internal Medicine, Department of Gastroenterology and Endocrinology, University of Marburg, Marburg, Germany Abstract. Transforming growth factor beta (TGF β) controls complex induces the canonical Smad signaling molecules which vital cellular functions through its ability to regulate gene then translocate into the nucleus to regulate transcription (2). The expression. TGFβ binding to its transmembrane receptor cellular response to TGF β can be extremely variable depending kinases initiates distinct intracellular signalling cascades on the cell type and the activation status of a cell at a given time. including the Smad signalling and transcription factors and also For instance, TGF β induces growth arrest and apoptosis in Smad-independent pathways. In normal epithelial cells, TGF β healthy epithelial cells, whereas it can also promote tumor stimulation induces a cytostatic program which includes the progression through stimulation of cell proliferation and the transcriptional repression of the c-Myc oncogene and the later induction of an epithelial-to-mesenchymal transition of tumor induction of the cell cycle inhibitors p15 INK4b and p21 Cip1 . cells (1, 3). In the last decade it has become clear that both the During carcinogenesis, however, many tumor cells lose their tumor suppressing and the tumor promoting functions of TGF β ability to respond to TGF β with growth inhibition, and instead, are primarily regulated on the level of gene expression through activate genes involved in cell proliferation, invasion and Smad-dependent and -independent mechanisms (1, 2, 4). -

Supplementary Materials
Supplementary Materials + - NUMB E2F2 PCBP2 CDKN1B MTOR AKT3 HOXA9 HNRNPA1 HNRNPA2B1 HNRNPA2B1 HNRNPK HNRNPA3 PCBP2 AICDA FLT3 SLAMF1 BIC CD34 TAL1 SPI1 GATA1 CD48 PIK3CG RUNX1 PIK3CD SLAMF1 CDKN2B CDKN2A CD34 RUNX1 E2F3 KMT2A RUNX1 T MIXL1 +++ +++ ++++ ++++ +++ 0 0 0 0 hematopoietic potential H1 H1 PB7 PB6 PB6 PB6.1 PB6.1 PB12.1 PB12.1 Figure S1. Unsupervised hierarchical clustering of hPSC-derived EBs according to the mRNA expression of hematopoietic lineage genes (microarray analysis). Hematopoietic-competent cells (H1, PB6.1, PB7) were separated from hematopoietic-deficient ones (PB6, PB12.1). In this experiment, all hPSCs were tested in duplicate, except PB7. Genes under-expressed or over-expressed in blood-deficient hPSCs are indicated in blue and red respectively (related to Table S1). 1 C) Mesoderm B) Endoderm + - KDR HAND1 GATA6 MEF2C DKK1 MSX1 GATA4 WNT3A GATA4 COL2A1 HNF1B ZFPM2 A) Ectoderm GATA4 GATA4 GSC GATA4 T ISL1 NCAM1 FOXH1 NCAM1 MESP1 CER1 WNT3A MIXL1 GATA4 PAX6 CDX2 T PAX6 SOX17 HBB NES GATA6 WT1 SOX1 FN1 ACTC1 ZIC1 FOXA2 MYF5 ZIC1 CXCR4 TBX5 PAX6 NCAM1 TBX20 PAX6 KRT18 DDX4 TUBB3 EPCAM TBX5 SOX2 KRT18 NKX2-5 NES AFP COL1A1 +++ +++ 0 0 0 0 ++++ +++ ++++ +++ +++ ++++ +++ ++++ 0 0 0 0 +++ +++ ++++ +++ ++++ 0 0 0 0 hematopoietic potential H1 H1 H1 H1 H1 H1 PB6 PB6 PB7 PB7 PB6 PB6 PB7 PB6 PB6 PB6.1 PB6.1 PB6.1 PB6.1 PB6.1 PB6.1 PB12.1 PB12.1 PB12.1 PB12.1 PB12.1 PB12.1 Figure S2. Unsupervised hierarchical clustering of hPSC-derived EBs according to the mRNA expression of germ layer differentiation genes (microarray analysis) Selected ectoderm (A), endoderm (B) and mesoderm (C) related genes differentially expressed between hematopoietic-competent (H1, PB6.1, PB7) and -deficient cells (PB6, PB12.1) are shown (related to Table S1). -

Supplemental Table 1. Primers and Probes for RT-Pcrs
Supplemental Table 1. Primers and probes for RT-PCRs Gene Direction Sequence Quantitative RT-PCR E2F1 Forward Primer 5’-GGA TTT CAC ACC TTT TCC TGG AT-3’ Reverse Primer 5’-CCT GGA AAC TGA CCA TCA GTA CCT-3’ Probe 5’-FAM-CGA GCT GGC CCA CTG CTC TCG-TAMRA-3' E2F2 Forward Primer 5'-TCC CAA TCC CCT CCA GAT C-3' Reverse Primer 5'-CAA GTT GTG CGA TGC CTG C-3' Probe 5' -FAM-TCC TTT TGG CCG GCA GCC G-TAMRA-3' E2F3a Forward Primer 5’-TTT AAA CCA TCT GAG AGG TAC TGA TGA-3’ Reverse Primer 5’-CGG CCC TCC GGC AA-3’ Probe 5’-FAM-CGC TTT CTC CTA GCT CCA GCC TTC G-TAMRA-3’ E2F3b Forward Primer 5’-TTT AAA CCA TCT GAG AGG TAC TGA TGA-3’ Reverse Primer 5’-CCC TTA CAG CAG CAG GCA A-3’ Probe 5’-FAM-CGC TTT CTC CTA GCT CCA GCC TTC G-TAMRA-3’ IRF-1 Forward Primer 5’-TTT GTA TCG GCC TGT GTG AAT G-3’ Reverse Primer 5’-AAG CAT GGC TGG GAC ATC A-3’ Probe 5’-FAM-CAG CTC CGG AAC AAA CAG GCA TCC TT-TAMRA-3' IRF-2 Forward Primer 5'-CGC CCC TCG GCA CTC T-3' Reverse Primer 5'-TCT TCC TAT GCA GAA AGC GAA AC-3' Probe 5'-FAM-TTC ATC GCT GGG CAC ACT ATC AGT-TAMRA-3' TBP Forward Primer 5’-CAC GAA CCA CGG CAC TGA TT-3’ Reverse Primer 5’-TTT TCT TGC TGC CAG TCT GGA C-3’ Probe 5’-FAM-TGT GCA CAG GAG CCA AGA GTG AAG A-BHQ1-3’ Primers and Probes for quantitative RT-PCRs were designed using the computer program “Primer Express” (Applied Biosystems, Foster City, CA, USA). -

Mammalian Atypical E2fs Link Endocycle Control to Cancer
Mammalian Atypical E2Fs Link Endocycle Control to Cancer DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Hui-Zi Chen Graduate Program in Integrated Biomedical Science Program The Ohio State University 2011 Dissertation Committee: Gustavo Leone, PhD, Advisor Michael Ostrowski, PhD Clay Marsh, MD Tsonwin Hai, PhD Kathryn Wikenheiser-Brokamp, MD PhD Copyright by Hui-Zi Chen 2011 Abstract The endocycle is a developmentally programmed variant cell cycle consisting of successive S (DNA synthesis) and G (Gap) phases without an intervening M phase or cytokinesis. As a consequence of the regulated “decoupling” of DNA replication and mitosis, which are two central processes of the traditional cell division program, endocycling cells acquire highly polyploid genomes after having undergone multiple rounds of whole genome reduplication. Although essential for metazoan development, relatively little is known about the regulation of endocycle or its physiologic role in higher vertebrates such as the mammal. A substantial body of work in the model organism Drosophila melanogaster has demonstrated an important function for dE2Fs in the control of endocycle. Genetic studies showed that both endocycle initiation and progression is severely disrupted by altering the expression of the fly E2F activator (dE2F1) or repressor (dE2F2). In mammals, the E2F family is comprised of nine structurally related proteins, encoded by eight distinct genes, that can be classified into transcriptional activators (E2f1, E2f2, E2f3a and E2f3b) or repressors (E2f4, E2f5, E2f6, E2f7 and E2f8). The repressor subclass may then be further divided into canonical (E2f4, E2f5 and E2f6) or atypical E2fs (E2f7 and E2f8). -

Forced Activation of Notch in Macrophages Represses Tumor Growth By
Author Manuscript Published OnlineFirst on January 12, 2016; DOI: 10.1158/0008-5472.CAN-15-2019 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. Forced Activation of Notch in Macrophages Represses Tumor Growth by Upregulating miR-125a and Disabling Tumor-Associated Macrophages Jun-Long Zhao1, Fei Huang1, Fei He2, Chun-Chen Gao1, Shi-Qian Liang1, Peng-Fei Ma2, Guang-Ying Dong1, Hua Han1, 2, Hong-Yan Qin1 1State Key Laboratory of Cancer Biology, Department of Medical Genetics and Developmental Biology, 2Department of Hepatic Surgery, Xijing Hospital, Fourth Military Medical University, Xi’an 710032, China Corresponding Authors: Hong-Yan Qin: Department of Medical Genetics and Developmental Biology, Fourth Military Medical University, Chang-Le Xi Street #169, Xi’an 710032, China; Tel.: +86 29 84774487; Fax: +86 29 83246270; Email: [email protected] Hua Han: Department of Medical Genetics and Developmental Biology, Fourth Military Medical University, Chang-Le Xi Street #169, Xi’an 710032, China; Tel.: +86 29 84774513, Fax: +86 29 83246270; Email: [email protected] Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/). J-L. Zhao and F. Huang contributed equally to this study. Running title: Notch regulates TAMs through miR-125a Disclosure of Potential Conflicts of Interest: The authors disclose no potential conflicts of interest. 1 Downloaded from cancerres.aacrjournals.org on September 28, 2021. © 2016 American Association for Cancer Research. Author Manuscript Published OnlineFirst on January 12, 2016; DOI: 10.1158/0008-5472.CAN-15-2019 Author manuscripts have been peer reviewed and accepted for publication but have not yet been edited. -

Evolutionarily Conserved Anterior Expansion of the Central Nervous System Promoted by a Common Pcg-Hox Program
© 2018. Published by The Company of Biologists Ltd | Development (2018) 145, dev160747. doi:10.1242/dev.160747 RESEARCH ARTICLE Evolutionarily conserved anterior expansion of the central nervous system promoted by a common PcG-Hox program Behzad Yaghmaeian Salmani, Ignacio Monedero Cobeta, Jonathan Rakar, Susanne Bauer, Jesús Rodriguez Curt, Annika Starkenberg and Stefan Thor* ABSTRACT A regulatory program well-suited for contributing to the A conserved feature of the central nervous system (CNS) is the establishment of anterior CNS expansion is the Polycomb group prominent expansion of anterior regions (brain) compared with (PcG) complex and the Hox homeotic genes. The PcG complex is a posterior (nerve cord). The cellular and regulatory processes driving collective name for several subcomplexes; Polycomb repressor anterior CNS expansion are not well understood in any bilaterian complexes 1 and 2 (PRC1 and PRC2) have received most attention species. Here, we address this expansion in Drosophila and mouse. (Steffen and Ringrose, 2014; Piunti and Shilatifard, 2016). These We find that, compared with the nerve cord, the brain displays complexes act to modify key residues in histone tails, with PRC1 extended progenitor proliferation, more elaborate daughter cell ubiquitylating H2A and PRC2 methylating H3K27, resulting in proliferation and more rapid cell cycle speed in both Drosophila and transcriptional repression of target genes. Among the best-known mouse. These features contribute to anterior CNS expansion in both PcG target genes are the Hox homeotic genes, which encode species. With respect to genetic control, enhanced brain proliferation clustered arrays of related transcription factors that play key roles in is severely reduced by ectopic Hox gene expression, by either Hox anterior-posterior (A-P) patterning of the body plan, including the misexpression or by loss of Polycomb group (PcG) function. -
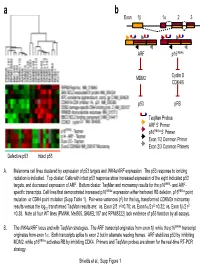
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S.