Bio-Plex Manager Software 6.2 User Guide
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Eminem 1 Eminem
Eminem 1 Eminem Eminem Eminem performing live at the DJ Hero Party in Los Angeles, June 1, 2009 Background information Birth name Marshall Bruce Mathers III Born October 17, 1972 Saint Joseph, Missouri, U.S. Origin Warren, Michigan, U.S. Genres Hip hop Occupations Rapper Record producer Actor Songwriter Years active 1995–present Labels Interscope, Aftermath Associated acts Dr. Dre, D12, Royce da 5'9", 50 Cent, Obie Trice Website [www.eminem.com www.eminem.com] Marshall Bruce Mathers III (born October 17, 1972),[1] better known by his stage name Eminem, is an American rapper, record producer, and actor. Eminem quickly gained popularity in 1999 with his major-label debut album, The Slim Shady LP, which won a Grammy Award for Best Rap Album. The following album, The Marshall Mathers LP, became the fastest-selling solo album in United States history.[2] It brought Eminem increased popularity, including his own record label, Shady Records, and brought his group project, D12, to mainstream recognition. The Marshall Mathers LP and his third album, The Eminem Show, also won Grammy Awards, making Eminem the first artist to win Best Rap Album for three consecutive LPs. He then won the award again in 2010 for his album Relapse and in 2011 for his album Recovery, giving him a total of 13 Grammys in his career. In 2003, he won the Academy Award for Best Original Song for "Lose Yourself" from the film, 8 Mile, in which he also played the lead. "Lose Yourself" would go on to become the longest running No. 1 hip hop single.[3] Eminem then went on hiatus after touring in 2005. -

Lady Sovereign Tvs Last Thursday Night to Wit- Ness the Passing of “ER” to What I’M Sure Will Be a Golden Reign in Alex Terrono Diversify Her Album, the Short Reruns
END OF AN ‘ER’(A) CAMPUS AWAY FOR BANDS THE SUMMMER? Rock out on campus and check out Eve Samborn covers moving out of town some of our very own Wash. U. talent for the summer in Forum today. in Scene. INSIDE Miss the season finale PAGE 6 PAGE 5 of “ER”? Check out Marcia McIntosh’s recap in Ca- denza. BACK PAGE Sthe independentTUDENT newspaper of Washington University in St. Louis LIFE since eighteen seventy-eightg h Vol. 130 No. 76 www.studlife.com Wednesday, April 8, 2009 STDs at WU University to fi nish construction more common than perceived on South 40 before fall move-in William Shim will also offer the South 40 a large nience store,” Carroll wrote. experience and provide basic ameni- Becca Krock Staff Reporter multipurpose room for all types of Jeremy Lai, a sophomore who ties to make living on campus more Staff Reporter uses,” Carroll wrote in an e-mail. will be the student technology coor- convenient for students. STD numbers Wohl’s residential area will form dinator for the new Wohl residential Despite such new perks of liv- After a construction period of a residential college along with area, said there are several benefi ts ing on the South 40, Thompson still The statistical prevalence of across the U.S. over a full school year, the new Rubelmann Hall and new Umrath to living there. decided to live in the Village next sexually transmitted infections on HPV 6.2 million Wohl Center and the new Umrath Hall. “I will never have to leave my year. -

Double Play.Puz
Double Play Pete Muller ACROSS 1 2 3 4 5 6 7 8 9 10 11 12 13 1. "When in ___" (song by Billy Joel 14 15 16 or Nickel Creek) 5. "Too Pooped ___" (Chuck Berry 17 18 19 tune) 10. Improvisation à la Ella 20 21 22 14. Contest judged by Steven Tyler for two seasons, familiarly 23 24 25 15. Sushi bar fish often coated with 26 27 28 29 30 31 32 33 teriyaki 16. Aces, say 34 35 36 37 17. "Three Times a Lady" band 19. Short introduction of a subject? 38 39 40 41 20. Duke who said "If music be the food of love, play on" 42 43 44 45 21. Brad Paisley/Carrie Underwood 46 47 48 49 duet that starts "We didn't care if people stared" 50 51 52 53 23. Djoko, e.g. 25. All of Latin? 54 55 56 57 58 59 60 26. Bush court appointee who describes himself as a "practical 61 62 63 originalist" 64 65 66 29. Prominent object on Boston's first album cover 67 68 69 32. Director Almodóvar 34. Joan Baez's singing sister ©2017 | The meta for this puzzle is a Jimmy Buffett song that would complete this puzzle's theme entries. 37. Jakob Dylan vocal quality 38. Neurotransmitter studied in org. 66. Seaweed's sister in "Hairspray" 22. Derisive nickname for Tyrion chem. 67. Wall St. deals that use a lot of Lannister on "Game of Thrones," 39. It's full of unknowns borrowed money with "the" 41. -
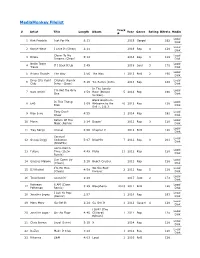
Mediamonkey Filelist
MediaMonkey Filelist Track # Artist Title Length Album Year Genre Rating Bitrate Media # Local 1 Kirk Franklin Just For Me 5:11 2019 Gospel 182 Disk Local 2 Kanye West I Love It (Clean) 2:11 2019 Rap 4 128 Disk Closer To My Local 3 Drake 5:14 2014 Rap 3 128 Dreams (Clean) Disk Nellie Tager Local 4 If I Back It Up 3:49 2018 Soul 3 172 Travis Disk Local 5 Ariana Grande The Way 3:56 The Way 1 2013 RnB 2 190 Disk Drop City Yacht Crickets (Remix Local 6 5:16 T.I. Remix (Intro 2013 Rap 128 Club Intro - Clean) Disk In The Lonely I'm Not the Only Local 7 Sam Smith 3:59 Hour (Deluxe 5 2014 Pop 190 One Disk Version) Block Brochure: In This Thang Local 8 E40 3:09 Welcome to the 16 2012 Rap 128 Breh Disk Soil 1, 2 & 3 They Don't Local 9 Rico Love 4:55 1 2014 Rap 182 Know Disk Return Of The Local 10 Mann 3:34 Buzzin' 2011 Rap 3 128 Macc (Remix) Disk Local 11 Trey Songz Unusal 4:00 Chapter V 2012 RnB 128 Disk Sensual Local 12 Snoop Dogg Seduction 5:07 BlissMix 7 2012 Rap 0 201 Disk (BlissMix) Same Damn Local 13 Future Time (Clean 4:49 Pluto 11 2012 Rap 128 Disk Remix) Sun Come Up Local 14 Glasses Malone 3:20 Beach Cruiser 2011 Rap 128 (Clean) Disk I'm On One We the Best Local 15 DJ Khaled 4:59 2 2011 Rap 5 128 (Clean) Forever Disk Local 16 Tessellated Searchin' 2:29 2017 Jazz 2 173 Disk Rahsaan 6 AM (Clean Local 17 3:29 Bleuphoria 2813 2011 RnB 128 Patterson Remix) Disk I Luh Ya Papi Local 18 Jennifer Lopez 2:57 1 2014 Rap 193 (Remix) Disk Local 19 Mary Mary Go Get It 2:24 Go Get It 1 2012 Gospel 4 128 Disk LOVE? [The Local 20 Jennifer Lopez On the -

2021Equine 'Health' and 'Care' Catalog
■ Bio Harnesses ■ Harness Repairs ■ Equine Health Products ■ Divine Equine Products ■ Training Equipment ■ Horse Blankets ■ Bowman Bits ■ Nutra-GloTM ■ Say Whoa!® ■ VetWrap ■ Tingley Boots & Raincoats ■ Eberglo Your ONE STOP for everything harnesses and more! 2021 Equine 'Health' and 'Care' Catalog 74 S. Little Britain Rd. Nottingham, PA 19362 717-529-7506 Animal Health Products Welcome to our 2021 Retail Catalog! To Our Valued Customer; First and foremost we want to thank you for your business and for giving us the opportunity to serve you and your horses' needs. We continue to add new items to our already large inventory found in this catalog. If you need a product not found in our catalog just ask, we can most likely get it for you. To explore our complete lineup of products including the Animal Health Products, Harnesses and Halters, visit our store in Nottingham some time. We appreciate your feedback and any suggestions or questions you may have regarding our products and services. We are here to help! Aaron King This is only a partial catalog (#1 of 3). We are currently working on (#2 of 3) with our full line of harness and halters, (#3 of 3) will Windy Knoll Harness include our training and sales prep harnesses. 74 S. Little Britain Rd. Disclaimer: Prices in the catalog are subject to change. If there Nottingham, PA 19362 is a significant increase we will notify you before shipping your order. Product images in this catalog are not actual size. Please Phone: 717-529-7506 read container size and weight of all products. -

Culture and Contempt: the Limitations of Expressive Criminal Law
Culture and Contempt: The Limitations of Expressive Criminal Law Ted Sampsell-Jones* The law is the master teacher and guides each generation as to what is acceptable conduct. - Asa Hutchinson' The law of the land in America is full of shit. - Chuck D' I. INTRODUCTION Over the past decade, legal scholars have paid increasing attention to ways that criminal law affects social norms and socialization. While these ideas are not entirely original,1 the renewed focus on criminal law's role in social construction has been illuminating nonetheless. The recent scholarship has reminded us that criminal laws prevent crime not only by applying legal sanctions ' The author received an A.B. from Dartmouth, a J.D. from Yale Law School, and is currently clerking for Judge William Fletcher on the Ninth Circuit Court of Appeals. Thanks to Michelle Drake, Bob Ellickson, Elizabeth Emens, Owen Fiss, David Fontana, Bernard Harcourt, Neal Katyal, Heather Lewis, Richard McAdams, Brian Nelson, and Sara Sampsell-Jones for their suggestions and comments. Asa Hutchinson, Administrator, U.S. Drug Enforcement Admin., Debate with Gov. Gary Johnson (N.M.) at the Yale Law School (Nov. 15, 2001), available at http://www.usdoj.gov/dea/speeches/sl 11501.html. 'CHUCK D, FIGHT THE POWER: RAP, RACE, AND REALITY 14 (1997). 1. See Mark Tushnet, Everything Old Is New Again, 1998 WISC. L. REV. 579. By tracing the ideological development of the "new" school of criminal law scholarship, Bernard Harcourt has questioned its originality. BERNARD E. HARCOURT, ILLUSION OF ORDER: THE FALSE PROMISE OF BROKEN WINDOWS POLICING 1-16, 24-56 (2001). -

Bio-Plex Pro Assays
…… ………. NEW magnetic protocol and software requirements! Bio-Plex Pro™ Assays Cytokine, Chemokine, and Growth Factors Instruction Manual For technical support, call your local Bio-Rad office, or in the U.S., call 1-800-4BIORAD (1-800-424-6723). For research use only. Not for diagnostic procedures. Table of Contents Section 1 Introduction 1 Section 2 Principle 3 Section 3 Product Description 5 Section 4 Recommended Materials 8 Section 5 Bead Regions 9 Section 6 Sample Preparation 10 Section 7 Standard Preparation 12 Section 8 Assay Instructions 16 8.1 Assay Plate and Wash Format 16 8.2 Plan Experiment 16 8.3 Calibrate Vacuum Apparatus 17 8.4 Prepare Coupled Magnetic Beads 18 8.5 Assay Procedure 19 Section 9 Data Acquisition 23 Section 10 Troubleshooting Guide 29 Section 11 Safety Considerations 35 Section 12 Plate Layout Template 36 Section 13 Legal Notice 38 Section 1 Introduction Cytokines, chemokines, and growth factors are cell signaling proteins, mediating a wide range of physiological responses, including immunity, inflammation, and hematopoiesis. They are also associated with a spectrum of diseases ranging from tumor growth to infections to Parkinson’s disease. These molecules are typically measured either by bioassay or immunoassay. Both techniques are time consuming and can facilitate the analysis of only a single target at a time. The Bio-Plex® system, which incorporates novel technology using color-coded beads, permits the simultaneous detection of up to 100 cytokines in a single well of a 96-well microplate. Bio-Plex Pro™ cytokine, chemokine, and growth factor assays are magnetic bead-based multiplex assays designed to measure multiple cytokines, chemokines, and growth factors in diverse matrices like serum, plasma, and tissue culture supernatants. -

Confessions of a Hip-Hop Hippie
California State University, San Bernardino CSUSB ScholarWorks Electronic Theses, Projects, and Dissertations Office of aduateGr Studies 6-2014 CONFESSIONS OF A HIP-HOP HIPPIE Tristan D. Acker California State University, San Bernardino, CA 92407 Follow this and additional works at: https://scholarworks.lib.csusb.edu/etd Part of the Poetry Commons Recommended Citation Acker, Tristan D., "CONFESSIONS OF A HIP-HOP HIPPIE" (2014). Electronic Theses, Projects, and Dissertations. 60. https://scholarworks.lib.csusb.edu/etd/60 This Thesis is brought to you for free and open access by the Office of aduateGr Studies at CSUSB ScholarWorks. It has been accepted for inclusion in Electronic Theses, Projects, and Dissertations by an authorized administrator of CSUSB ScholarWorks. For more information, please contact [email protected]. CONFESSIONS OF A HIP-HOP HIPPIE THE MEASURE OF AN EMPTY CAR A Project Presented to the Faculty of California State University, San Bernardino In Partial Fulfillment of the Requirements for the Degree Master of Fine Arts in Creative Writing: Poetry by Tristan Douglas Acker June 2014 CONFESSIONS OF A HIP-HOP HIPPIE THE MEASURE OF AN EMPTY CAR A Project Presented to the Faculty of California State University, San Bernardino by Tristan Douglas Acker June 2014 Approved by: John Chad Sweeney, First Reader Juan Delgado, Second Reader © 2014 Tristan Douglas Acker ABSTRACT This Statement of Purpose does not give a history of hip-hop or hip-hop poetry but rather how this particular poet fits into the current phase of hip-hop and performance poetry. In it, I discuss and explain the new pro-working class hip-hop performance poetic. -

Liste Collection En Vente CD Vinyl Hip Hop Funk Soul Crunk Gfunk Jazz Rnb HIP HOP MUSIC MUSEUM PRIX Négociable En Lot
1 Liste collection en vente CD Vinyl Hip Hop Funk Soul crunk GFunk Jazz Rnb HIP HOP MUSIC MUSEUM PRIX négociable en lot Type Prix Nom de l’artiste Nom de l’album de Genre Année Etat Qté média + de hits disco funk 2CD Funk 2001 1 12 2pac How do u want it CD Single Hip Hop 1996 1 5,50 2pac Ru still down 2CD Hip Hop 1997 1 14 2pac Until the end of time CD Single Hip Hop 2001 1 1,30 2pac Until the end of time CD Hip Hop 2001 0 5,99 2 pac Loyal to the game CD Hip Hop 2004 1 4,99 2 Pac Part 1 THUG CD Digi Hip Hop 2007 1 12 50 Cent Get Rich or Die Trying CD Hip Hop 2003 1 2,90 50 CENT The new breed CD DVD Hip Hop 2003 1 6 50 Cent The Massacre CD Hip Hop 2005 1 1,90 50 Cent The Massacre CD dvd Hip Hop 2005 0 7,99 50 Cent Curtis CD Hip Hop 2007 0 3,50 50 Cent After Curtis CD Diggi Hip Hop 2007 1 4 50 Cent Self District CD Hip Hop 2009 0 3,50 100% Ragga Dj Ewone CD 2005 1 7 Reggaetton 504 BOYZ BALLERS CD Hip Hop 2002 US 1 6 Aaliyah One In A Million CD Hip Hop 1996 1 14,99 Aaliyah I refuse & more than a CD single RNB 2001 1 8 woman Aaliyah feat Timbaland We need a resolution CD Single RNB 2001 1 5,60 2 Aaliyah More than a woman Vinyl Hip HOP 2001 1 5 Aaliyah CD Hip Hop 2001 2 4,99 Aaliyah Don’t know what to tell ya CD Single RNB 2002 1 4,30 Aaliyah I care for you 2CD RNB 2002 1 11 AG The dirty version CD Hip Hop 1999 Neuf 1 4 A motown special VINYL Funk 1977 1 3,50 disco album Akon Konvikt mixtape vol 2 CD Digi Hip Hop 1 4 Akon trouble CD Hip hop 2004 1 4,20 Akon Konvicted CD Hip Hop 2006 1 1,99 Akon Freedom CD Hip Hop 2008 1 2 Al green I cant stop CD Jazz 2003 1 1,90 Alicia Keys Songs in a minor (coffret CD Single Hip Hop 2001 1 25 Taiwan) album TAIWAN Maxi Alici Keys The Diary of CD Hip Hop 2004 1 4,95 Allstars deejay’s Mix dj crazy b dee nasty 2CD MIX 2002 1 3,90 eanov pone lbr Alyson Williams CD RNB 1992 1 3,99 Archie Bell Tightening it up CD Funk 1994 Us 10 6,50 Artmature Hip Hop VOL 2 CD Hip Hop 2015 1 19 Art Porter Pocket city CD Funk 1992 1 7 Ashanti CD RNB 2002 1 6 Asher Roth I love college VINYL Hip Hop 2009 1 8,99 A.S.M. -

Tupac Shakur 29 Dr
EXPONENTES DEL VERSO computación PDF generado usando el kit de herramientas de fuente abierta mwlib. Ver http://code.pediapress.com/ para mayor información. PDF generated at: Sat, 08 Mar 2014 05:48:13 UTC Contenidos Artículos Capitulo l: El mundo de el HIP HOP 1 Hip hop 1 Capitulo ll: Exponentes del HIP HOP 22 The Notorious B.I.G. 22 Tupac Shakur 29 Dr. Dre 45 Snoop Dogg 53 Ice Cube 64 Eminem 72 Nate Dogg 86 Cartel de Santa 90 Referencias Fuentes y contribuyentes del artículo 93 Fuentes de imagen, Licencias y contribuyentes 95 Licencias de artículos Licencia 96 1 Capitulo l: El mundo de el HIP HOP Hip hop Hip Hop Orígenes musicales Funk, Disco, Dub, R&B, Soul, Toasting, Doo Wop, scat, Blues, Jazz Orígenes culturales Años 1970 en el Bronx, Nueva York Instrumentos comunes Tocadiscos, Sintetizador, DAW, Caja de ritmos, Sampler, Beatboxing, Guitarra, bajo, Piano, Batería, Violin, Popularidad 1970 : Costa Este -1973 : Costa, Este y Oeste - 1980 : Norte America - 1987 : Países Occidentales - 1992 : Actualidad - Mundial Derivados Electro, Breakbeat, Jungle/Drum and Bass, Trip Hop, Grime Subgéneros Rap Alternativo, Gospel Hip Hop, Conscious Hip Hop, Freestyle Rap, Gangsta Rap, Hardcore Hip Hop, Horrorcore, nerdcore hip hop, Chicano rap, jerkin', Hip Hop Latinoamericano, Hip Hop Europeo, Hip Hop Asiatico, Hip Hop Africano Fusiones Country rap, Hip Hop Soul, Hip House, Crunk, Jazz Rap, MerenRap, Neo Soul, Nu metal, Ragga Hip Hop, Rap Rock, Rap metal, Hip Life, Low Bap, Glitch Hop, New Jack Swing, Electro Hop Escenas regionales East Coast, West Coast, -

NOT in OUR HOUSE Black History Month XU Asks Itself, "Where Do We Go from Here?" by SARAH KELLEY There, It Was Very Motivational
Xavier University Exhibit All Xavier Student Newspapers Xavier Student Newspapers 2000-02-02 Xavier University Newswire Xavier University (Cincinnati, Ohio) Follow this and additional works at: https://www.exhibit.xavier.edu/student_newspaper Recommended Citation Xavier University (Cincinnati, Ohio), "Xavier University Newswire" (2000). All Xavier Student Newspapers. 2844. https://www.exhibit.xavier.edu/student_newspaper/2844 This Book is brought to you for free and open access by the Xavier Student Newspapers at Exhibit. It has been accepted for inclusion in All Xavier Student Newspapers by an authorized administrator of Exhibit. For more information, please contact [email protected]. .U -N IVE RS . I TY 85thyear, issue 17 week of FEBRUARY 2,-2000 www.xu.edu/soahiewswire/ Campus kick~ off NOT IN OUR HOUSE Black History Month XU asks itself, "Where do we go from here?" BY SARAH KELLEY there, it was very motivational. I'm Senior News Editor ready to go out and celebrate." Last night, a candlelight vigil In the closing remarks of the - kicked off Xavier's annual celebra ceremony, Director of Multicultural tion of-Black History Month in an Affairs Mila Cooper expressed the opening ceremony sponsored by need to "celebrate the hope we have the Office of Multicultural Affairs. for the future, eradic'ate the hate that Seven candles were lit to repre still exists and replace it with love sent black ancestors, the present! and understanding." the future, love, unity, courage and In the spirit of the themes intro respect This served as a reminder duced at the opening ceremony, a to focus on the past, present and variety of Xavier organizations are future while celebrating black his- sponsoring events throughout the tory. -
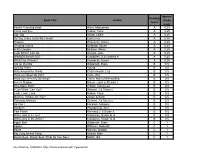
Book Title Author Reading Level Approx. Grade Level
Approx. Reading Book Title Author Grade Level Level Anno's Counting Book Anno, Mitsumasa A 0.25 Count and See Hoban, Tana A 0.25 Dig, Dig Wood, Leslie A 0.25 Do You Want To Be My Friend? Carle, Eric A 0.25 Flowers Hoenecke, Karen A 0.25 Growing Colors McMillan, Bruce A 0.25 In My Garden McLean, Moria A 0.25 Look What I Can Do Aruego, Jose A 0.25 What Do Insects Do? Canizares, S.& Chanko,P A 0.25 What Has Wheels? Hoenecke, Karen A 0.25 Cat on the Mat Wildsmith, Brain B 0.5 Getting There Young B 0.5 Hats Around the World Charlesworth, Liza B 0.5 Have you Seen My Cat? Carle, Eric B 0.5 Have you seen my Duckling? Tafuri, Nancy/Greenwillow B 0.5 Here's Skipper Salem, Llynn & Stewart,J B 0.5 How Many Fish? Cohen, Caron Lee B 0.5 I Can Write, Can You? Stewart, J & Salem,L B 0.5 Look, Look, Look Hoban, Tana B 0.5 Mommy, Where are You? Ziefert & Boon B 0.5 Runaway Monkey Stewart, J & Salem,L B 0.5 So Can I Facklam, Margery B 0.5 Sunburn Prokopchak, Ann B 0.5 Two Points Kennedy,J. & Eaton,A B 0.5 Who Lives in a Tree? Canizares, Susan et al B 0.5 Who Lives in the Arctic? Canizares, Susan et al B 0.5 Apple Bird Wildsmith, Brain C 1 Apples Williams, Deborah C 1 Bears Kalman, Bobbie C 1 Big Long Animal Song Artwell, Mike C 1 Brown Bear, Brown Bear What Do You See? Martin, Bill C 1 Found online, 7/20/2012, http://home.comcast.net/~ngiansante/ Approx.