Quantification and Confirmation of Fifteen Carbamate Pesticide
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Modifications on the Basic Skeletons of Vinblastine and Vincristine
Molecules 2012, 17, 5893-5914; doi:10.3390/molecules17055893 OPEN ACCESS molecules ISSN 1420-3049 www.mdpi.com/journal/molecules Review Modifications on the Basic Skeletons of Vinblastine and Vincristine Péter Keglevich, László Hazai, György Kalaus and Csaba Szántay * Department of Organic Chemistry and Technology, University of Technology and Economics, H-1111 Budapest, Szt. Gellért tér 4, Hungary * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel: +36-1-463-1195; Fax: +36-1-463-3297. Received: 30 March 2012; in revised form: 9 May 2012 / Accepted: 10 May 2012 / Published: 18 May 2012 Abstract: The synthetic investigation of biologically active natural compounds serves two main purposes: (i) the total synthesis of alkaloids and their analogues; (ii) modification of the structures for producing more selective, more effective, or less toxic derivatives. In the chemistry of dimeric Vinca alkaloids enormous efforts have been directed towards synthesizing new derivatives of the antitumor agents vinblastine and vincristine so as to obtain novel compounds with improved therapeutic properties. Keywords: antitumor therapy; vinblastine; vincristine; derivatives 1. Introduction Vinblastine (1) and vincristine (2) are dimeric alkaloids (Figure 1) isolated from the Madagaskar periwinkle plant (Catharantus roseus), exhibit significant cytotoxic activity and are used in the antitumor therapy as antineoplastic agents. In the course of cell proliferation they act as inhibitors during the metaphase of the cell cycle and by binding to the microtubules inhibit the development of the mitotic spindle. In tumor cells these agents inhibit the DNA repair and the RNA synthesis mechanisms, blocking the DNA-dependent RNA polymerase. Molecules 2012, 17 5894 Figure 1. -

Carbamate Pesticides Aldicarb Aldicarb Sulfoxide Aldicarb Sulfone
Connecticut General Statutes Sec 19a-29a requires the Commissioner of Public Health to annually publish a list setting forth all analytes and matrices for which certification for testing is required. Connecticut ELCP Drinking Water Analytes Revised 05/31/2018 Microbiology Total Coliforms Fecal Coliforms/ E. Coli Carbamate Pesticides Legionella Aldicarb Cryptosporidium Aldicarb Sulfoxide Giardia Aldicarb Sulfone Carbaryl Physicals Carbofuran Turbidity 3-Hydroxycarbofuran pH Methomyl Conductivity Oxamyl (Vydate) Minerals Chlorinated Herbicides Alkalinity, as CaCO3 2,4-D Bromide Dalapon Chloride Dicamba Chlorine, free residual Dinoseb Chlorine, total residual Endothall Fluoride Picloram Hardness, Calcium as Pentachlorophenol CaCO3 Hardness, Total as CaCO3 Silica Chlorinated Pesticides/PCB's Sulfate Aldrin Chlordane (Technical) Nutrients Dieldrin Endrin Ammonia Heptachlor Nitrate Heptachlor Epoxide Nitrite Lindane (gamma-BHC) o-Phosphate Metolachlor Total Phosphorus Methoxychlor PCB's (individual aroclors) Note 1 PCB's (as decachlorobiphenyl) Note 1 Demands Toxaphene TOC Nitrogen-Phosphorus Compounds Alachlor Metals Atrazine Aluminum Butachlor Antimony Diquat Arsenic Glyphosate Barium Metribuzin Beryllium Paraquat Boron Propachlor Cadmium Simazine Calcium Chromium Copper SVOC's Iron Benzo(a)pyrene Lead bis-(2-ethylhexyl)phthalate Magnesium bis-(ethylhexyl)adipate Manganese Hexachlorobenzene Mercury Hexachlorocyclopentadiene Molybdenum Nickel Potassium Miscellaneous Organics Selenium Dibromochloropropane (DBCP) Silver Ethylene Dibromide (EDB) -

WO 2018/005077 Al O O© O
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/005077 Al 04 January 2018 (04.01.2018) W ! P O PCT (51) International Patent Classification: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, A61K 31/78 (2006.01) C08J 7/04 (2006.01) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. C08G 59/77 (2006.01) (84) Designated States (unless otherwise indicated, for every (21) International Application Number: kind of regional protection available): ARIPO (BW, GH, PCT/US20 17/037 176 GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, (22) International Filing Date: TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, 13 June 2017 (13.06.2017) EE, ES, FI, FR, GB, GR, HR, HU, IE, IS, IT, LT, LU, LV, (25) Filing Language: English MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, (26) Publication Language: English KM, ML, MR, NE, SN, TD, TG). (30) Priority Data: 62/356,918 30 June 2016 (30.06.2016) US Published: — with international search report (Art. 21(3)) (71) Applicant: ELEMENTIS SPECIALTIES, INC. [US/US]; 469 Old Trenton Road, East Windsor, NJ 085 12 (US). (72) Inventors: IJDO, Wouter; 1224 Bridle Estates Dri ve, Yardley, PA 19067 (US). CHEN, Yanhui; 4 Hal- stead Place, Princeton, NJ 08540 (US). -

Chemical Name Federal P Code CAS Registry Number Acutely
Acutely / Extremely Hazardous Waste List Federal P CAS Registry Acutely / Extremely Chemical Name Code Number Hazardous 4,7-Methano-1H-indene, 1,4,5,6,7,8,8-heptachloro-3a,4,7,7a-tetrahydro- P059 76-44-8 Acutely Hazardous 6,9-Methano-2,4,3-benzodioxathiepin, 6,7,8,9,10,10- hexachloro-1,5,5a,6,9,9a-hexahydro-, 3-oxide P050 115-29-7 Acutely Hazardous Methanimidamide, N,N-dimethyl-N'-[2-methyl-4-[[(methylamino)carbonyl]oxy]phenyl]- P197 17702-57-7 Acutely Hazardous 1-(o-Chlorophenyl)thiourea P026 5344-82-1 Acutely Hazardous 1-(o-Chlorophenyl)thiourea 5344-82-1 Extremely Hazardous 1,1,1-Trichloro-2, -bis(p-methoxyphenyl)ethane Extremely Hazardous 1,1a,2,2,3,3a,4,5,5,5a,5b,6-Dodecachlorooctahydro-1,3,4-metheno-1H-cyclobuta (cd) pentalene, Dechlorane Extremely Hazardous 1,1a,3,3a,4,5,5,5a,5b,6-Decachloro--octahydro-1,2,4-metheno-2H-cyclobuta (cd) pentalen-2- one, chlorecone Extremely Hazardous 1,1-Dimethylhydrazine 57-14-7 Extremely Hazardous 1,2,3,4,10,10-Hexachloro-6,7-epoxy-1,4,4,4a,5,6,7,8,8a-octahydro-1,4-endo-endo-5,8- dimethanonaph-thalene Extremely Hazardous 1,2,3-Propanetriol, trinitrate P081 55-63-0 Acutely Hazardous 1,2,3-Propanetriol, trinitrate 55-63-0 Extremely Hazardous 1,2,4,5,6,7,8,8-Octachloro-4,7-methano-3a,4,7,7a-tetra- hydro- indane Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]- 51-43-4 Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]-, P042 51-43-4 Acutely Hazardous 1,2-Dibromo-3-chloropropane 96-12-8 Extremely Hazardous 1,2-Propylenimine P067 75-55-8 Acutely Hazardous 1,2-Propylenimine 75-55-8 Extremely Hazardous 1,3,4,5,6,7,8,8-Octachloro-1,3,3a,4,7,7a-hexahydro-4,7-methanoisobenzofuran Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime 26419-73-8 Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime. -
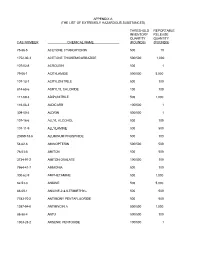
The List of Extremely Hazardous Substances)
APPENDIX A (THE LIST OF EXTREMELY HAZARDOUS SUBSTANCES) THRESHOLD REPORTABLE INVENTORY RELEASE QUANTITY QUANTITY CAS NUMBER CHEMICAL NAME (POUNDS) (POUNDS) 75-86-5 ACETONE CYANOHYDRIN 500 10 1752-30-3 ACETONE THIOSEMICARBAZIDE 500/500 1,000 107-02-8 ACROLEIN 500 1 79-06-1 ACRYLAMIDE 500/500 5,000 107-13-1 ACRYLONITRILE 500 100 814-68-6 ACRYLYL CHLORIDE 100 100 111-69-3 ADIPONITRILE 500 1,000 116-06-3 ALDICARB 100/500 1 309-00-2 ALDRIN 500/500 1 107-18-6 ALLYL ALCOHOL 500 100 107-11-9 ALLYLAMINE 500 500 20859-73-8 ALUMINUM PHOSPHIDE 500 100 54-62-6 AMINOPTERIN 500/500 500 78-53-5 AMITON 500 500 3734-97-2 AMITON OXALATE 100/500 100 7664-41-7 AMMONIA 500 100 300-62-9 AMPHETAMINE 500 1,000 62-53-3 ANILINE 500 5,000 88-05-1 ANILINE,2,4,6-TRIMETHYL- 500 500 7783-70-2 ANTIMONY PENTAFLUORIDE 500 500 1397-94-0 ANTIMYCIN A 500/500 1,000 86-88-4 ANTU 500/500 100 1303-28-2 ARSENIC PENTOXIDE 100/500 1 THRESHOLD REPORTABLE INVENTORY RELEASE QUANTITY QUANTITY CAS NUMBER CHEMICAL NAME (POUNDS) (POUNDS) 1327-53-3 ARSENOUS OXIDE 100/500 1 7784-34-1 ARSENOUS TRICHLORIDE 500 1 7784-42-1 ARSINE 100 100 2642-71-9 AZINPHOS-ETHYL 100/500 100 86-50-0 AZINPHOS-METHYL 10/500 1 98-87-3 BENZAL CHLORIDE 500 5,000 98-16-8 BENZENAMINE, 3-(TRIFLUOROMETHYL)- 500 500 100-14-1 BENZENE, 1-(CHLOROMETHYL)-4-NITRO- 500/500 500 98-05-5 BENZENEARSONIC ACID 10/500 10 3615-21-2 BENZIMIDAZOLE, 4,5-DICHLORO-2-(TRI- 500/500 500 FLUOROMETHYL)- 98-07-7 BENZOTRICHLORIDE 100 10 100-44-7 BENZYL CHLORIDE 500 100 140-29-4 BENZYL CYANIDE 500 500 15271-41-7 BICYCLO[2.2.1]HEPTANE-2-CARBONITRILE,5- -

N-Methyl Carbamate Cumulative Risk Assessment: Pilot Cumulative Analysis
UNITED STATES ENVIRONMENTAL PROTECTION AGENCY WASHINGTON, D.C. 20460 OFFICE OF PREVENTION, PESTICIDES, AND TOXIC SUBSTANCES April 15, 2005 MEMORANDUM SUBJECT: Transmittal of Meeting Minutes of the FIFRA Scientific Advisory Panel Meeting Held February 15 - 18, 2005 on the N-methyl Carbamate Cumulative Risk Assessment: Pilot Cumulative Analysis TO: James J. Jones, Director Office of Pesticide Programs FROM: Myrta R. Christian, Designated Federal Official Joseph E. Bailey, Designated Federal Official FIFRA Scientific Advisory Panel Office of Science Coordination and Policy THRU: Larry C. Dorsey, Executive Secretary FIFRA Scientific Advisory Panel Office of Science Coordination and Policy Clifford J. Gabriel, Director Office of Science Coordination and Policy Attached, please find the meeting minutes of the FIFRA Scientific Advisory Panel open meeting held in Arlington, Virginia on February 15 - 18, 2005. This report addresses a set of scientific issues being considered by the Environmental Protection Agency pertaining to the N- methyl carbamate cumulative risk assessment: pilot cumulative analysis. Attachment 1 of 113 cc: Susan Hazen Margaret Schneider Anne Lindsay Margie Fehrenbach Janet Andersen Debbie Edwards Steven Bradbury William Diamond Arnold Layne Tina Levine Lois Rossi Frank Sanders Richard Keigwin Randolph Perfetti William Jordan Douglas Parsons Enesta Jones Vanessa Vu (SAB) Anna Lowit David J. Miller Nelson Thurman Dirk Young David Hrdy Jeff Evans Steve Nako Stephanie Padilla R. Woodrow Setzer Ginger Moser Miles Okino Jerry Blancato Fred Power Curtis Dary Tom Nolan, USGS OPP Docket 2 of 113 FIFRA Scientific Advisory Panel Members Stephen M. Roberts, Ph.D. (Chair of the FIFRA SAP) Janice E. Chambers, Ph.D. H. Christopher Frey, Ph.D. -

Selective Effects of Carbamate Pesticides on Rat Neuronal Nicotinic Acetylcholine Receptors and Rat Brain Acetylcholinesterase Chantal J.G.M
Available online at www.sciencedirect.com R Toxicology and Applied Pharmacology 193 (2003) 139–146 www.elsevier.com/locate/taap Selective effects of carbamate pesticides on rat neuronal nicotinic acetylcholine receptors and rat brain acetylcholinesterase Chantal J.G.M. Smulders,a Tjerk J.H. Bueters,b Regina G.D.M. Van Kleef,a and Henk P.M. Vijverberga,* a Institute for Risk Assessment Sciences, Utrecht University, P.O. Box 80176, NL-3508 TD Utrecht, The Netherlands b TNO Prins Maurits Laboratory, P.O. Box 45, NL-2280 AA Rijswijk, The Netherlands Received 14 May 2003; accepted 31 July 2003 Abstract Effects of commonly used carbamate pesticides on rat neuronal nicotinic acetylcholine receptors expressed in Xenopus laevis oocytes have been investigated using the two-electrode voltage clamp technique. The potencies of these effects have been compared to the potencies of the carbamates to inhibit rat brain acetylcholinesterase. The potency order of six carbamates to inhibit ␣44 nicotinic receptors is fenoxycarb Ͼ EPTC Ͼ carbaryl, bendiocarb Ͼ propoxur Ͼ aldicarb with IC50 values ranging from 3 M for fenoxycarb to 165 M for propoxur and Ͼ1 mM for aldicarb. Conversely, the potency order of these carbamates to inhibit rat brain acetylcholinesterase is bendiocarb Ͼ propoxur, aldicarb Ͼ carbaryl ϾϾ EPTC, fenoxycarb with IC50 values ranging from 1 M for bendiocarb to 17 M for carbaryl and ϾϾ1 mM for EPTC and fenoxycarb. The ␣42, ␣34, and ␣32 nicotinic acetylcholine receptors are inhibited by fenoxycarb, EPTC, and carbaryl with potency orders similar to that for ␣44 receptors. Comparing the potencies of inhibition of the distinct subtypes of nicotinic acetylcholine receptors shows that the ␣32 receptor is less sensitive to inhibition by fenoxycarb and EPTC. -

Chapter 6 PRETREATMENT for NERVE AGENT EXPOSURE
Pretreatment for Nerve Agent Exposure Chapter 6 PRETREATMENT FOR NERVE AGENT EXPOSURE MICHAEL A. DUNN, M.D., FACP*; BRENNIE E. HACKLEY, JR., PH.D.†; AND FREDERICK R. SIDELL, M.D.‡ INTRODUCTION AGING OF NERVE AGENT–BOUND ACETYLCHOLINESTERASE PYRIDOSTIGMINE, A PERIPHERALLY ACTING CARBAMATE COMPOUND Efficacy Safety Wartime Use Improved Delivery CENTRALLY ACTING NERVE AGENT PRETREATMENTS NEW DIRECTIONS: BIOTECHNOLOGICAL PRETREATMENTS SUMMARY *Colonel, Medical Corps, U.S. Army; Director, Clinical Consultation, Office of the Assistant Secretary of Defense (Health Affairs), Washing- ton, D.C. 20301-1200; formerly, Commander, U.S. Army Medical Research Institute of Chemical Defense, Aberdeen Proving Ground, Mary- land 21010-5425 †Scientific Advisor, U.S. Army Medical Research Institute of Chemical Defense, Aberdeen Proving Ground, Maryland 21010-5425 ‡Formerly, Chief, Chemical Casualty Care Office, and Director, Medical Management of Chemical Casualties Course, U.S. Army Medical Research Institute of Chemical Defense, Aberdeen Proving Ground, Maryland 21010-5425; currently, Chemical Casualty Consultant, 14 Brooks Road, Bel Air, Maryland 21014 181 Medical Aspects of Chemical and Biological Warfare INTRODUCTION Nerve agents are rapidly acting chemical com- cal as well and may impair physical and mental pounds that can cause respiratory arrest within performance. A pretreatment must be administered minutes of absorption. Their speed of action im- to an entire force under a nerve agent threat. Any poses a need for rapid and appropriate reaction by resulting performance decrement, even a compara- exposed soldiers, their buddies, or medics, who tively minor one, would make pretreatment use must administer antidotes quickly enough to save unacceptable in battlefield situations requiring lives. A medical defense against nerve agents that maximum alertness and performance for survival. -

Code Chemical P026 1-(O-Chlorophenyl)Thiourea P081 1
Code Chemical P026 1-(o-Chlorophenyl)thiourea P081 1,2,3-Propanetriol, trinitrate (R) P042 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]-, (R)- P067 1,2-Propylenimine P185 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)- carbonyl]oxime 1,4,5,8-Dimethanonaphthalene, 1,2,3,4,10,10-hexa- chloro-1,4,4a,5,8,8a,-hexahydro-, P004 (1alpha,4alpha, 4abeta,5alpha,8alpha,8abeta)- 1,4,5,8-Dimethanonaphthalene, 1,2,3,4,10,10-hexa- chloro-1,4,4a,5,8,8a-hexahydro-, P060 (1alpha,4alpha, 4abeta,5beta,8beta,8abeta)- P002 1-Acetyl-2-thiourea P048 2,4-Dinitrophenol P051 2,7:3,6-Dimethanonaphth [2,3-b]oxirene, 3,4,5,6,9,9 -hexachloro-1a,2,2a,3,6,6a,7,7a- octahydro-, (1aalpha,2beta,2abeta,3alpha,6alpha,6abeta,7 beta, 7aalpha)-, & metabolites 2,7:3,6-Dimethanonaphth[2,3-b]oxirene, 3,4,5,6,9,9- hexachloro-1a,2,2a,3,6,6a,7,7a- P037 octahydro-, (1aalpha,2beta,2aalpha,3beta,6beta,6aalpha,7 beta, 7aalpha)- P045 2-Butanone, 3,3-dimethyl-1-(methylthio)-, O-[methylamino)carbonyl] oxime P034 2-Cyclohexyl-4,6-dinitrophenol 2H-1-Benzopyran-2-one, 4-hydroxy-3-(3-oxo-1- phenylbutyl)-, & salts, when present at P001 concentrations greater than 0.3% P069 2-Methyllactonitrile P017 2-Propanone, 1-bromo- P005 2-Propen-1-ol P003 2-Propenal P102 2-Propyn-1-ol P007 3(2H)-Isoxazolone, 5-(aminomethyl)- P027 3-Chloropropionitrile P047 4,6-Dinitro-o-cresol, & salts P059 4,7-Methano-1H-indene, 1,4,5,6,7,8,8-heptachloro- 3a,4,7,7a-tetrahydro- P008 4-Aminopyridine P008 4-Pyridinamine P007 5-(Aminomethyl)-3-isoxazolol 6,9-Methano-2,4,3-benzodioxathiepin, 6,7,8,9,10,10- -

Redalyc.Calidad Del Alcohol Producido En Europa Ilegalmente O
Adicciones ISSN: 0214-4840 [email protected] Sociedad Científica Española de Estudios sobre el Alcohol, el Alcoholismo y las otras Toxicomanías España Lachenmeier, Dirk W.; Leitz, Jenny; Schoeberl, Kerstin; Kuballa, Thomas; Straub, Irene; Rehm, Jürgen Calidad del alcohol producido en Europa ilegalmente o de forma no regulada: resultados del proyecto AMPHORA Adicciones, vol. 23, núm. 2, 2011, pp. 133-140 Sociedad Científica Española de Estudios sobre el Alcohol, el Alcoholismo y las otras Toxicomanías Palma de Mallorca, España Disponible en: http://www.redalyc.org/articulo.oa?id=289122828006 Cómo citar el artículo Número completo Sistema de Información Científica Más información del artículo Red de Revistas Científicas de América Latina, el Caribe, España y Portugal Página de la revista en redalyc.org Proyecto académico sin fines de lucro, desarrollado bajo la iniciativa de acceso abierto Calidad del alcohol producido en Europa ilegalmente o de forma no regulada: resultados del proyecto AMPHORA Quality of illegally and informally produced alcohol in Europe: Results from the AMPHORA project Dirk W. Lachenmeier*; Jenny Leitz*; * Chemisches und Veterinäruntersuchungsamt (CVUA) Karlsruhe, Germany kerstin schoeberL*; thomas kubaLLa*; ** Centre for Addiction and Mental Health (CAMH), Toronto, Canada irene straub*; Jürgen rehm**,***,**** *** Dalla Lana School of Public Health, University of Toronto, Toronto, Canada **** Epidemiological Research Unit, Institute for Clinical Psychology and Psychotherapy, TU Dresden, Dresden, Germany Enviar correspondencia a: Dirk W. Lachenmeier. Chemisches und Veterinäruntersuchungsamt (CVUA) Karlsruhe, Weissenburger Strasse 3, D-76187 Karlsruhe, Germany. Tel: +49-721-926-5434, Fax: +49-721-926-5539 Email: [email protected]. recibido: septiembre 2010 aceptado: enero 2011 RESUMEN ABSTRACT Antecedentes. En la región europea de la OMS, el consumo promedio Background. -

A Simple and Efficient Green Method for the Deprotection of N-Boc in Various Structurally Diverse Amines Under Water-Mediated Catalyst-Free Conditions
International Journal of Chemistry; Vol. 4, No. 3; 2012 ISSN 1916-9698 E-ISSN 1916-9701 Published by Canadian Center of Science and Education A Simple and Efficient Green Method for the Deprotection of N-Boc in Various Structurally Diverse Amines under Water-mediated Catalyst-free Conditions Cheraiet Zinelaabidine1, Ouarna Souad1, Jamel Zoubir1, Berredjem Malika1 & Aouf Nour-Eddine1 1 LCOA, Bioorganic Chemistry Group, Sciences Faculty, Chemistry Department, Badji Mokhtar-Annaba University, Algeria Correspondence: Aouf Nour-Eddine, LCOA, Bioorganic Chemistry Group, Sciences Faculty, Chemistry Department, Badji Mokhtar-Annaba University, BP 12, 23000, Algeria. Tel: 213-3887-2789. E-mail: [email protected] Received: February 8, 2012 Accepted: February 27, 2012 Published: May 27, 2012 doi:10.5539/ijc.v4n3p73 URL: http://dx.doi.org/10.5539/ijc.v4n3p73 This work was generously supported by the (Direction Generale de la Recherche Scientifique et du Développement Technologique, DGRS-DT), Algerian Ministry of Scientific Research, (FNR), and fruitful discussions with Dr. Malika Ibrahim-Ouali, Université d’Aix Marseille III, France were greatly appreciated Abstract A simple, efficient and eco-friendly protocol has been developed for the deprotection of N-Boc on structurally diverse amines. Selective removal of N-Boc groups was achieved with excellent yields using water around reflux temperatures. In the absence of any additional reagents, this method represents a reasonable alternative to previously reported deprotection procedures. Keywords: boc, deprotection, water, green chemistry, amines, cyclosulfamides 1. Introduction The development of a simple and effective method, using an environmentally friendly approach as well as an economical process is in great demand in protective group chemistry. -

Recent Advances on Detection of Insecticides Using Optical Sensors
sensors Review Recent Advances on Detection of Insecticides Using Optical Sensors Nurul Illya Muhamad Fauzi 1, Yap Wing Fen 1,2,*, Nur Alia Sheh Omar 1,2 and Hazwani Suhaila Hashim 2 1 Functional Devices Laboratory, Institute of Advanced Technology, Universiti Putra Malaysia, Serdang 43400, Selangor, Malaysia; [email protected] (N.I.M.F.); [email protected] (N.A.S.O.) 2 Department of Physics, Faculty of Science, Universiti Putra Malaysia, Serdang 43400, Selangor, Malaysia; [email protected] * Correspondence: [email protected] Abstract: Insecticides are enormously important to industry requirements and market demands in agriculture. Despite their usefulness, these insecticides can pose a dangerous risk to the safety of food, environment and all living things through various mechanisms of action. Concern about the environmental impact of repeated use of insecticides has prompted many researchers to develop rapid, economical, uncomplicated and user-friendly analytical method for the detection of insecticides. In this regards, optical sensors are considered as favorable methods for insecticides analysis because of their special features including rapid detection time, low cost, easy to use and high selectivity and sensitivity. In this review, current progresses of incorporation between recognition elements and optical sensors for insecticide detection are discussed and evaluated well, by categorizing it based on insecticide chemical classes, including the range of detection and limit of detection. Additionally, this review aims to provide powerful insights to researchers for the future development of optical sensors in the detection of insecticides. Citation: Fauzi, N.I.M.; Fen, Y.W.; Omar, N.A.S.; Hashim, H.S. Recent Keywords: insecticides; optical sensor; recognition element Advances on Detection of Insecticides Using Optical Sensors.