Genetic Variability of Hosta Virus X in Hosta
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Abacca Mosaic Virus
Annex Decree of Ministry of Agriculture Number : 51/Permentan/KR.010/9/2015 date : 23 September 2015 Plant Quarantine Pest List A. Plant Quarantine Pest List (KATEGORY A1) I. SERANGGA (INSECTS) NAMA ILMIAH/ SINONIM/ KLASIFIKASI/ NAMA MEDIA DAERAH SEBAR/ UMUM/ GOLONGA INANG/ No PEMBAWA/ GEOGRAPHICAL SCIENTIFIC NAME/ N/ GROUP HOST PATHWAY DISTRIBUTION SYNONIM/ TAXON/ COMMON NAME 1. Acraea acerata Hew.; II Convolvulus arvensis, Ipomoea leaf, stem Africa: Angola, Benin, Lepidoptera: Nymphalidae; aquatica, Ipomoea triloba, Botswana, Burundi, sweet potato butterfly Merremiae bracteata, Cameroon, Congo, DR Congo, Merremia pacifica,Merremia Ethiopia, Ghana, Guinea, peltata, Merremia umbellata, Kenya, Ivory Coast, Liberia, Ipomoea batatas (ubi jalar, Mozambique, Namibia, Nigeria, sweet potato) Rwanda, Sierra Leone, Sudan, Tanzania, Togo. Uganda, Zambia 2. Ac rocinus longimanus II Artocarpus, Artocarpus stem, America: Barbados, Honduras, Linnaeus; Coleoptera: integra, Moraceae, branches, Guyana, Trinidad,Costa Rica, Cerambycidae; Herlequin Broussonetia kazinoki, Ficus litter Mexico, Brazil beetle, jack-tree borer elastica 3. Aetherastis circulata II Hevea brasiliensis (karet, stem, leaf, Asia: India Meyrick; Lepidoptera: rubber tree) seedling Yponomeutidae; bark feeding caterpillar 1 4. Agrilus mali Matsumura; II Malus domestica (apel, apple) buds, stem, Asia: China, Korea DPR (North Coleoptera: Buprestidae; seedling, Korea), Republic of Korea apple borer, apple rhizome (South Korea) buprestid Europe: Russia 5. Agrilus planipennis II Fraxinus americana, -

Grapevine Virus Diseases: Economic Impact and Current Advances in Viral Prospection and Management1
1/22 ISSN 0100-2945 http://dx.doi.org/10.1590/0100-29452017411 GRAPEVINE VIRUS DISEASES: ECONOMIC IMPACT AND CURRENT ADVANCES IN VIRAL PROSPECTION AND MANAGEMENT1 MARCOS FERNANDO BASSO2, THOR VINÍCIUS MArtins FAJARDO3, PASQUALE SALDARELLI4 ABSTRACT-Grapevine (Vitis spp.) is a major vegetative propagated fruit crop with high socioeconomic importance worldwide. It is susceptible to several graft-transmitted agents that cause several diseases and substantial crop losses, reducing fruit quality and plant vigor, and shorten the longevity of vines. The vegetative propagation and frequent exchanges of propagative material among countries contribute to spread these pathogens, favoring the emergence of complex diseases. Its perennial life cycle further accelerates the mixing and introduction of several viral agents into a single plant. Currently, approximately 65 viruses belonging to different families have been reported infecting grapevines, but not all cause economically relevant diseases. The grapevine leafroll, rugose wood complex, leaf degeneration and fleck diseases are the four main disorders having worldwide economic importance. In addition, new viral species and strains have been identified and associated with economically important constraints to grape production. In Brazilian vineyards, eighteen viruses, three viroids and two virus-like diseases had already their occurrence reported and were molecularly characterized. Here, we review the current knowledge of these viruses, report advances in their diagnosis and prospection of new species, and give indications about the management of the associated grapevine diseases. Index terms: Vegetative propagation, plant viruses, crop losses, berry quality, next-generation sequencing. VIROSES EM VIDEIRAS: IMPACTO ECONÔMICO E RECENTES AVANÇOS NA PROSPECÇÃO DE VÍRUS E MANEJO DAS DOENÇAS DE ORIGEM VIRAL RESUMO-A videira (Vitis spp.) é propagada vegetativamente e considerada uma das principais culturas frutíferas por sua importância socioeconômica mundial. -

Downloaded in July 2020
viruses Article The Phylogeography of Potato Virus X Shows the Fingerprints of Its Human Vector Segundo Fuentes 1, Adrian J. Gibbs 2 , Mohammad Hajizadeh 3, Ana Perez 1 , Ian P. Adams 4, Cesar E. Fribourg 5, Jan Kreuze 1 , Adrian Fox 4 , Neil Boonham 6 and Roger A. C. Jones 7,* 1 Crop and System Sciences Division, International Potato Center, La Molina Lima 15023, Peru; [email protected] (S.F.); [email protected] (A.P.); [email protected] (J.K.) 2 Emeritus Faculty, Australian National University, Canberra, ACT 2600, Australia; [email protected] 3 Plant Protection Department, Faculty of Agriculture, University of Kurdistan, Sanandaj 6617715175, Iran; [email protected] 4 Fera Science Ltd., Sand Hutton York YO41 1LZ, UK; [email protected] (I.P.A.); [email protected] (A.F.) 5 Departamento de Fitopatologia, Universidad Nacional Agraria, La Molina Lima 12056, Peru; [email protected] 6 Institute for Agrifood Research Innovations, Newcastle University, Newcastle upon Tyne NE1 7RU, UK; [email protected] 7 UWA Institute of Agriculture, University of Western Australia, 35 Stirling Highway, Crawley, WA 6009, Australia * Correspondence: [email protected] Abstract: Potato virus X (PVX) occurs worldwide and causes an important potato disease. Complete PVX genomes were obtained from 326 new isolates from Peru, which is within the potato crop0s main Citation: Fuentes, S.; Gibbs, A.J.; domestication center, 10 from historical PVX isolates from the Andes (Bolivia, Peru) or Europe (UK), Hajizadeh, M.; Perez, A.; Adams, I.P.; and three from Africa (Burundi). Concatenated open reading frames (ORFs) from these genomes Fribourg, C.E.; Kreuze, J.; Fox, A.; plus 49 published genomic sequences were analyzed. -
(A) Journals with the Largest Number of Papers Reporting Estimates Of
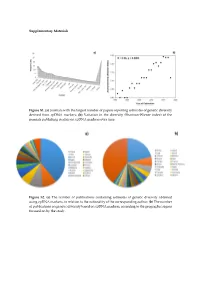
Supplementary Materials Figure S1. (a) Journals with the largest number of papers reporting estimates of genetic diversity derived from cpDNA markers; (b) Variation in the diversity (Shannon-Wiener index) of the journals publishing studies on cpDNA markers over time. Figure S2. (a) The number of publications containing estimates of genetic diversity obtained using cpDNA markers, in relation to the nationality of the corresponding author; (b) The number of publications on genetic diversity based on cpDNA markers, according to the geographic region focused on by the study. Figure S3. Classification of the angiosperm species investigated in the papers that analyzed genetic diversity using cpDNA markers: (a) Life mode; (b) Habitat specialization; (c) Geographic distribution; (d) Reproductive cycle; (e) Type of flower, and (f) Type of pollinator. Table S1. Plant species identified in the publications containing estimates of genetic diversity obtained from the use of cpDNA sequences as molecular markers. Group Family Species Algae Gigartinaceae Mazzaella laminarioides Angiospermae Typhaceae Typha laxmannii Angiospermae Typhaceae Typha orientalis Angiospermae Typhaceae Typha angustifolia Angiospermae Typhaceae Typha latifolia Angiospermae Araliaceae Eleutherococcus sessiliflowerus Angiospermae Polygonaceae Atraphaxis bracteata Angiospermae Plumbaginaceae Armeria pungens Angiospermae Aristolochiaceae Aristolochia kaempferi Angiospermae Polygonaceae Atraphaxis compacta Angiospermae Apocynaceae Lagochilus macrodontus Angiospermae Polygonaceae Atraphaxis -

H. Kiyosumiensis F
Hosta Species Update●The Hosta Library © W. George Schmid 2006 H. kiyosumiensis F. Maekawa 1935 Jornal of Japanese Botany 11:689; ic. f. 15 1935 キヨスミギボウシ = Kyosumi Giboshi H. kiyosumiensis var. petrophila F. Maekawa 1938 Divisiones et Plantae Novae Generis Hostae (2). J. Japanese Botany, 14:1:45–49. イワマ ギボウシ = Iwama Giboshi History and Nomenclature: In Japan this species is called Kyiosumi Giboshi, the “Kiyosumi (Mountain) Hosta.” The species epithet stems from the Latinized name of Kiyosumiyama (清澄山), a small mountain (380 m/about 1250 feet AMSL with a great view of Tokyo Bay) located on Boso Peninsula (Bōsō-hantō; 房総半島), which is located in south Chiba Prefecture (Chiba-ken; 千葉県) forming the eastern edge of Tokyo Bay. Maekawa established H. kiyosumiensis in 1935 and published further on the species in 1938, when he also described a new variety of the species as H. (kiyosumiensis var.) petrophila. This varietal epithet is derived from the Latin “liking (or) growing on rocks.” The Japanese name Iwama Giboshi (イワマ ギボウシ) has the same meaning. The latter is differentiated by minor morphological details caused by its different, rocky habitat in Yamashiro province (山城国; Yamashiro-no kuni), H. kiyosumiensis Maekawa 1935 (in situ) Otogawa (男川) River; Okazaki-shi (岡崎市; formerly Nukata-cho) Aichi Prefecture (愛知県; Aichi-ken) 1 an old province of Japan (today the southern part of Kyoto Prefecture). Maekawa (1938) gave a much abbreviated Latin description and the variety is here considered synonymous with the species. In fact, Maekawa (1969) no longer supported the varietal rank. N. Fujita (1976) confirmed H. kiyosumiensis as a species and added two morphs previously described by Maekawa (1940), i.e., H. -

Viral Diversity in Tree Species
Universidade de Brasília Instituto de Ciências Biológicas Departamento de Fitopatologia Programa de Pós-Graduação em Biologia Microbiana Doctoral Thesis Viral diversity in tree species FLÁVIA MILENE BARROS NERY Brasília - DF, 2020 FLÁVIA MILENE BARROS NERY Viral diversity in tree species Thesis presented to the University of Brasília as a partial requirement for obtaining the title of Doctor in Microbiology by the Post - Graduate Program in Microbiology. Advisor Dra. Rita de Cássia Pereira Carvalho Co-advisor Dr. Fernando Lucas Melo BRASÍLIA, DF - BRAZIL FICHA CATALOGRÁFICA NERY, F.M.B Viral diversity in tree species Flávia Milene Barros Nery Brasília, 2025 Pages number: 126 Doctoral Thesis - Programa de Pós-Graduação em Biologia Microbiana, Universidade de Brasília, DF. I - Virus, tree species, metagenomics, High-throughput sequencing II - Universidade de Brasília, PPBM/ IB III - Viral diversity in tree species A minha mãe Ruth Ao meu noivo Neil Dedico Agradecimentos A Deus, gratidão por tudo e por ter me dado uma família e amigos que me amam e me apoiam em todas as minhas escolhas. Minha mãe Ruth e meu noivo Neil por todo o apoio e cuidado durante os momentos mais difíceis que enfrentei durante minha jornada. Aos meus irmãos André, Diego e meu sobrinho Bruno Kawai, gratidão. Aos meus amigos de longa data Rafaelle, Evanessa, Chênia, Tati, Leo, Suzi, Camilets, Ricardito, Jorgito e Diego, saudade da nossa amizade e dos bons tempos. Amo vocês com todo o meu coração! Minha orientadora e grande amiga Profa Rita de Cássia Pereira Carvalho, a quem escolhi e fui escolhida para amar e fazer parte da família. -

Reducing Deer Damage in Landscapes Part 2
Reducing Deer Damage in the Landscape D Coetzee Public Domain bestfriendthemom Dusty Hancock CC BY-NC-ND 2.0 Chatham Master Gardener Volunteer Matt Jones Horticulture Agent NC Cooperative Extension - Chatham County Center Plant Selection Deer Candy • Aucuba • Hosta • Arborvitae • Indian Hawthorn • Azalea • Ivy • Blueberry • Chionanthus, • Clematis Malus, Prunus, • Daylily Pyrus • Euonymus • Redbuds • Fatsia • Roses Muhenbergia capillaris Pink Muhly Grass (Poaceae) Andrea Laine Jim Robbins CC BY NC 4.0 CC BY-NC-ND 4.0 Muhlenbergia capillaris Pink Muhly Grass (Poaceae) Full Sun Moist to very dry 1-3’ x 1-3’ Fall Susan Strine Fall-Winter CC BY 2.0 Schizachyrium scoparium Little Blue Stem (Poaceae) Jim Robbins Joshua Mayer Jim Robbins Montreais CC BY-SA 2.0 DE CC BY-NC-ND 4.0 CC BY-NC-ND 4.0 CC BY-SA 3.0 Schizachyrium scoparium Little Blue Stem (Poaceae) Full Sun Moist to dry Good drainage 1-4’ x 18”-2’ Summer-Fall Susan Strine Summer-Fall Jim RobbinsCC BY 2.0 CC BY-NC-ND 4.0 Chasmanthium latifolium River Oats (Poaceae) Klasse im Garten Anne McCormack CC BY 2.0 CC BY-NC 2.0 Chasmanthium latifolium River Oats (Poaceae) Part shade to dappled sun Moist Occasionally wet 2-5’ x 1-2’ Summer-Fall Anne McCormack Summer-Fall CC BY-NC 2.0 Myrica cerifera (Myricaceae) Common Wax Myrtle Forest and Kim Starr CC BY 2.0 Forest and Kim Starr CC BY 2.0 Myrica cerifera (Myricaceae) Common Wax Myrtle Jim Robbins CC BY-NC-ND 4.0 Jim Robbins Jim Robbins CC BY-NC-ND 4.0 CC BY-NC-ND 4.0 Great for urban soils, full sun to part shade. -

Jan Dvorak (As Quickly Written Down by a Person with Poor Hearing…Me)
1/22/2018 The Impact of Polyploidy on Genome Evolution in Poales and Other Monocots “I don’t have to emphasize that gene duplications are the fabric of evolution in plants.” -Jan Dvorak (as quickly written down by a person with poor hearing…me) Michael R. McKain The University of Alabama @mrmckain @mrmckain Poales Diversity Grass genomes: the choose your own adventure of genome evolution • ~22,800 species • ~11,088 species in Poaceae • Transposons (McClintock, Wessler) • GC content bias (Carels and Bernardi 2000) • Three WGD events 0 0 4 0 0 • rho (Peterson et al. 2004) 3 y c n 0 e 0 u 2 q e r F • 0 sigma (Tang et al. 2010) 0 1 • tau (Tang et al. 2010, Jiao et al. 2014) 0 %GC Givnish et al. 2010 @mrmckain Schnable et al., 2009 Zeroing in on WGD placement Banana genome Pineapple genome How has ancient polyploidy altered the genomic landscape in grasses and other Poales? D’Hont et al. 2012 Ming, VanBuren et al. 2015 Recovered sigma after grass divergence from commelinids Recovered sigma after grass+pineapple divergence from commelinids @mrmckain @mrmckain 1 1/22/2018 Phylotranscriptomic approach Coalescence-based Phylogeny of 234 Single-copy genes • Sampling 27 transcriptomes and 7 genomes • Phylogeny consistent with previous • Representation for all families (except Thurniaceae) in nuclear gene results Poales • Conflicting topology with • RNA from young leaf or apical meristem, a combination of chloroplast genome tree: Moncot Tree of Life and 1KP • Ecdeiocolea/Joinvillea sister instead of • General steps: a grade • Trinity assembly • Typha -

Impact of Dietary Diversification on Invasive Slugs and Biological Control with Notes on Slug Species of Kentucky
University of Kentucky UKnowledge University of Kentucky Master's Theses Graduate School 2010 IMPACT OF DIETARY DIVERSIFICATION ON INVASIVE SLUGS AND BIOLOGICAL CONTROL WITH NOTES ON SLUG SPECIES OF KENTUCKY Anna K. Thomas University of Kentucky, [email protected] Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Thomas, Anna K., "IMPACT OF DIETARY DIVERSIFICATION ON INVASIVE SLUGS AND BIOLOGICAL CONTROL WITH NOTES ON SLUG SPECIES OF KENTUCKY" (2010). University of Kentucky Master's Theses. 35. https://uknowledge.uky.edu/gradschool_theses/35 This Thesis is brought to you for free and open access by the Graduate School at UKnowledge. It has been accepted for inclusion in University of Kentucky Master's Theses by an authorized administrator of UKnowledge. For more information, please contact [email protected]. ABSTRACT OF THESIS IMPACT OF DIETARY DIVERSIFICATION ON INVASIVE SLUGS AND BIOLOGICAL CONTROL WITH NOTES ON SLUG SPECIES OF KENTUCKY Increasing introductions of non-native terrestrial slugs (Mollusca: Gastropoda) are a concern to North American regulatory agencies as these generalists impact the yield and reduce the aesthetic value of crop plants. Understanding how the increase in diversification in North American cropping systems affects non-native gastropods and finding effective biological control options are imperative for pest management; however, little research has been done in this area. This study tested the hypothesis that dietary diversification affects the biological control capacity of a generalist predator and allows the slug pest Deroceras reticulatum (Müller) (Stylommatophora: Agriolimacidae) to more effectively fulfill its nutritional requirements. -

Molecular Characterization of a New Monopartite Dsrna Mycovirus from Mycorrhizal Thelephora Terrestris
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Virology 489 (2016) 12–19 Contents lists available at ScienceDirect Virology journal homepage: www.elsevier.com/locate/yviro Molecular characterization of a new monopartite dsRNA mycovirus from mycorrhizal Thelephora terrestris (Ehrh.) and its detection in soil oribatid mites (Acari: Oribatida) Karel Petrzik a,n, Tatiana Sarkisova a, Josef Starý b, Igor Koloniuk a, Lenka Hrabáková a,c, Olga Kubešová a a Department of Plant Virology, Institute of Plant Molecular Biology, Biology Centre of the Czech Academy of Sciences, Branišovská 31, 370 05 České Budějovice, Czech Republic b Institute of Soil Biology, Biology Centre of the Czech Academy of Sciences, Na Sádkách 7, 370 05 České Budějovice, Czech Republic c Department of Genetics, Faculty of Science, University of South Bohemia in České Budějovice, Branišovská 31a, 370 05 České Budějovice, Czech Republic article info abstract Article history: A novel dsRNA virus was identified in the mycorrhizal fungus Thelephora terrestris (Ehrh.) and sequenced. Received 28 July 2015 This virus, named Thelephora terrestris virus 1 (TtV1), contains two reading frames in different frames Returned to author for revisions but with the possibility that ORF2 could be translated as a fusion polyprotein after ribosomal -1 fra- 4 November 2015 meshifting. Picornavirus 2A-like motif, nudix hydrolase, phytoreovirus S7, and RdRp domains were found Accepted 10 November 2015 in a unique arrangement on the polyprotein. A new genus named Phlegivirus and containing TtV1, PgLV1, Available online 14 December 2015 RfV1 and LeV is therefore proposed. Twenty species of oribatid mites were identified in soil material in Keywords: the vicinity of T. -

ICTV Code Assigned: 2011.001Ag Officers)
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2011.001aG officers) Short title: Change existing virus species names to non-Latinized binomials (e.g. 6 new species in the genus Zetavirus) Modules attached 1 2 3 4 5 (modules 1 and 9 are required) 6 7 8 9 Author(s) with e-mail address(es) of the proposer: Van Regenmortel Marc, [email protected] Burke Donald, [email protected] Calisher Charles, [email protected] Dietzgen Ralf, [email protected] Fauquet Claude, [email protected] Ghabrial Said, [email protected] Jahrling Peter, [email protected] Johnson Karl, [email protected] Holbrook Michael, [email protected] Horzinek Marian, [email protected] Keil Guenther, [email protected] Kuhn Jens, [email protected] Mahy Brian, [email protected] Martelli Giovanni, [email protected] Pringle Craig, [email protected] Rybicki Ed, [email protected] Skern Tim, [email protected] Tesh Robert, [email protected] Wahl-Jensen Victoria, [email protected] Walker Peter, [email protected] Weaver Scott, [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp . -

Exploring the Tymovirids Landscape Through Metatranscriptomics Data
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.15.452586; this version posted July 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Exploring the tymovirids landscape through metatranscriptomics data 2 Nicolás Bejerman1,2, Humberto Debat1,2 3 4 1 Instituto de Patología Vegetal – Centro de Investigaciones Agropecuarias – Instituto Nacional de 5 Tecnología Agropecuaria (IPAVE-CIAP-INTA), Camino 60 Cuadras Km 5,5 (X5020ICA), Córdoba, 6 Argentina 7 2 Consejo Nacional de Investigaciones Científicas y Técnicas. Unidad de Fitopatología y Modelización 8 Agrícola, Camino 60 Cuadras Km 5,5 (X5020ICA), Córdoba, Argentina 9 10 Corresponding author: Nicolás Bejerman, [email protected] 11 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.15.452586; this version posted July 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 12 Abstract 13 Tymovirales is an order of viruses with positive-sense, single-stranded RNA genomes that mostly infect 14 plants, but also fungi and insects. The number of tymovirid sequences has been growing in the last few 15 years with the extensive use of high-throughput sequencing platforms. Here we report the discovery of 31 16 novel tymovirid genomes associated with 27 different host plant species, which were hidden in public 17 databases.