(Mygalomorphae, Atracinae), with Implications for Venom
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Interactions of Insecticidal Spider Peptide Neurotoxins with Insect Voltage- and Neurotransmitter-Gated Ion Channels
Interactions of insecticidal spider peptide neurotoxins with insect voltage- and neurotransmitter-gated ion channels (Molecular representation of - HXTX-Hv1c including key binding residues, adapted from Gunning et al, 2008) PhD Thesis Monique J. Windley UTS 2012 CERTIFICATE OF AUTHORSHIP/ORIGINALITY I certify that the work in this thesis has not previously been submitted for a degree nor has it been submitted as part of requirements for a degree except as fully acknowledged within the text. I also certify that the thesis has been written by me. Any help that I have received in my research work and the preparation of the thesis itself has been acknowledged. In addition, I certify that all information sources and literature used are indicated in the thesis. Monique J. Windley 2012 ii ACKNOWLEDGEMENTS There are many people who I would like to thank for contributions made towards the completion of this thesis. Firstly, I would like to thank my supervisor Prof. Graham Nicholson for his guidance and persistence throughout this project. I would like to acknowledge his invaluable advice, encouragement and his neverending determination to find a solution to any problem. He has been a valuable mentor and has contributed immensely to the success of this project. Next I would like to thank everyone at UTS who assisted in the advancement of this research. Firstly, I would like to acknowledge Phil Laurance for his assistance in the repair and modification of laboratory equipment. To all the laboratory and technical staff, particulary Harry Simpson and Stan Yiu for the restoration and sourcing of equipment - thankyou. I would like to thank Dr Mike Johnson for his continual assistance, advice and cheerful disposition. -

Non-Theraphosidae Mygalomorphs of North America
Non-Theraphosidae Mygalomorphs of North America © 2015 by Andrew Olson North America has a rich collection of fauna in all the families of Mygalomorphae. This paper introduces these families, with a special emphasis on the taxa of the USA. To see what nonTheraphosidae mygalomorphs appear in your state, please consult this large table: http://cacoseraph.x10host.com/ntmygs/ntmygs_usa.php Taxonomical placement of mygalomorph families found in the USA. Family arrangement based on Platnick’s World Spider Catalog and is indicative of the “closeness” of relation between various families. Kingdom: Animalia . Convenience taxonomical level: Invertebrata # . Phylum: Arthropoda . Class: Arachnida . Order: Araneae . Suborder: Opisthothelae . Infraorder: Mygalomorphae . Family: Atypidae / Atypidae Pursewebs or Atypical tarantulas “Atypical” ! . Family: Antrodiaetidae / Antrodiaetidae Folding trapdoors ! . Family: Mecicobothriidae / Mecicobothriidae Dwarf tarantulas ! . Family: Dipluridae / Dipluridae Funnelweb tarantulas “Two Tails” ! . Family: Cyrtaucheniidae / Cyrtaucheniidae Waferlid/Wafer trapdoors ! . Family: Ctenizidae / Ctenizidae Corklid trapdoors/Trapdoors . Family: Euctenizidae / Euctenizidae ????????? . Family: Nemesiidae / Nemesiidae False tarantulas . Family: Barychelidae / Barychelidae Brushed trapdoors/Brushfoot trapdoors ! . Family: Theraphosidae / Theraphosidae ** Tarantulas Key: WSC Link -

Miranda ZA 2018.Pdf
Zoologischer Anzeiger 273 (2018) 33–55 Contents lists available at ScienceDirect Zoologischer Anzeiger jou rnal homepage: www.elsevier.com/locate/jcz Review of Trichodamon Mello-Leitão 1935 and phylogenetic ଝ placement of the genus in Phrynichidae (Arachnida, Amblypygi) a,b,c,∗ a Gustavo Silva de Miranda , Adriano Brilhante Kury , a,d Alessandro Ponce de Leão Giupponi a Laboratório de Aracnologia, Museu Nacional do Rio de Janeiro, Universidade Federal do Rio de Janeiro, Quinta da Boa Vista s/n, São Cristóvão, Rio de Janeiro-RJ, CEP 20940-040, Brazil b Entomology Department, National Museum of Natural History, Smithsonian Institution, 10th St. & Constitution Ave NW, Washington, DC, 20560, USA c Center for Macroecology, Evolution and Climate, Natural History Museum of Denmark (Zoological Museum), University of Copenhagen, Universitetsparken 15, 2100, Copenhagen, Denmark d Servic¸ o de Referência Nacional em Vetores das Riquetsioses (LIRN), Colec¸ ão de Artrópodes Vetores Ápteros de Importância em Saúde das Comunidades (CAVAISC), IOC-FIOCRUZ, Manguinhos, 21040360, Rio de Janeiro, RJ, Brazil a r t i c l e i n f o a b s t r a c t Article history: Amblypygi Thorell, 1883 has five families, of which Phrynichidae is one of the most diverse and with a Received 18 October 2017 wide geographic distribution. The genera of this family inhabit mostly Africa, India and Southeast Asia, Received in revised form 27 February 2018 with one genus known from the Neotropics, Trichodamon Mello-Leitão, 1935. Trichodamon has two valid Accepted 28 February 2018 species, T. princeps Mello-Leitão, 1935 and T. froesi Mello-Leitão, 1940 which are found in Brazil, in the Available online 10 March 2018 states of Bahia, Goiás, Minas Gerais and Rio Grande do Norte. -

Tarantulas and Social Spiders
Tarantulas and Social Spiders: A Tale of Sex and Silk by Jonathan Bull BSc (Hons) MSc ICL Thesis Presented to the Institute of Biology of The University of Nottingham in Partial Fulfilment of the Requirements for the Degree of Doctor of Philosophy The University of Nottingham May 2012 DEDICATION To my parents… …because they both said to dedicate it to the other… I dedicate it to both ii ACKNOWLEDGEMENTS First and foremost I would like to thank my supervisor Dr Sara Goodacre for her guidance and support. I am also hugely endebted to Dr Keith Spriggs who became my mentor in the field of RNA and without whom my understanding of the field would have been but a fraction of what it is now. Particular thanks go to Professor John Brookfield, an expert in the field of biological statistics and data retrieval. Likewise with Dr Susan Liddell for her proteomics assistance, a truly remarkable individual on par with Professor Brookfield in being able to simplify even the most complex techniques and analyses. Finally, I would really like to thank Janet Beccaloni for her time and resources at the Natural History Museum, London, permitting me access to the collections therein; ten years on and still a delight. Finally, amongst the greats, Alexander ‘Sasha’ Kondrashov… a true inspiration. I would also like to express my gratitude to those who, although may not have directly contributed, should not be forgotten due to their continued assistance and considerate nature: Dr Chris Wade (five straight hours of help was not uncommon!), Sue Buxton (direct to my bench creepy crawlies), Sheila Keeble (ventures and cleans where others dare not), Alice Young (read/checked my thesis and overcame her arachnophobia!) and all those in the Centre for Biomolecular Sciences. -

Phylogenomic Analysis and Revised Classification of Atypoid Mygalomorph Spiders (Araneae, Mygalomorphae), with Notes on Arachnid Ultraconserved Element Loci
Phylogenomic analysis and revised classification of atypoid mygalomorph spiders (Araneae, Mygalomorphae), with notes on arachnid ultraconserved element loci Marshal Hedin1, Shahan Derkarabetian1,2,3, Adan Alfaro1, Martín J. Ramírez4 and Jason E. Bond5 1 Department of Biology, San Diego State University, San Diego, CA, United States of America 2 Department of Biology, University of California, Riverside, Riverside, CA, United States of America 3 Department of Organismic and Evolutionary Biology, Museum of Comparative Zoology, Harvard University, Cambridge, MA, United States of America 4 Division of Arachnology, Museo Argentino de Ciencias Naturales ``Bernardino Rivadavia'', Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Buenos Aires, Argentina 5 Department of Entomology and Nematology, University of California, Davis, CA, United States of America ABSTRACT The atypoid mygalomorphs include spiders from three described families that build a diverse array of entrance web constructs, including funnel-and-sheet webs, purse webs, trapdoors, turrets and silken collars. Molecular phylogenetic analyses have generally supported the monophyly of Atypoidea, but prior studies have not sampled all relevant taxa. Here we generated a dataset of ultraconserved element loci for all described atypoid genera, including taxa (Mecicobothrium and Hexurella) key to understanding familial monophyly, divergence times, and patterns of entrance web evolution. We show that the conserved regions of the arachnid UCE probe set target exons, such that it should be possible to combine UCE and transcriptome datasets in arachnids. We also show that different UCE probes sometimes target the same protein, and under the matching parameters used here show that UCE alignments sometimes include non- Submitted 1 February 2019 orthologs. Using multiple curated phylogenomic matrices we recover a monophyletic Accepted 28 March 2019 Published 3 May 2019 Atypoidea, and reveal that the family Mecicobothriidae comprises four separate and divergent lineages. -

Funnel Weaver Spiders (Funnel-Web Weavers, Grass Spiders)
Colorado Arachnids of Interest Funnel Weaver Spiders (Funnel-web weavers, Grass spiders) Class: Arachnida (Arachnids) Order: Araneae (Spiders) Family: Agelenidae (Funnel weaver Figure 1. Female grass spider on sheet web. spiders) Identification and Descriptive Features: Funnel weaver spiders are generally brownish or grayish spiders with a body typically ranging from1/3 to 2/3-inch when full grown. They have four pairs of eyes that are roughly the same size. The legs and body are hairy and legs usually have some dark banding. They are often mistaken for wolf spiders (Lycosidae family) but the size and pattern of eyes can most easily distinguish them. Like wolf spiders, the funnel weavers are very fast runners. Among the three most common genera (Agelenopsis, Hololena, Tegenaria) found in homes and around yards, Agelenopsis (Figures 1, 2 and 3) is perhaps most easily distinguished as it has long tail-like structures extending from the rear end of the body. These structures are the spider’s spinnerets, from which the silk emerges. Males of this genus have a unique and peculiarly coiled structure (embolus) on their pedipalps (Figure 3), the appendages next to the mouthparts. Hololena species often have similar appearance but lack the elongated spinnerets and male pedipalps have a normal clubbed appearance. Spiders within both genera Figure 2. Adult female of a grass spider, usually have dark longitudinal bands that run along the Agelenopsis sp. back of the cephalothorax and an elongated abdomen. Tegenaria species tend to have blunter abdomens marked with gray or black patches. Dark bands may also run along the cephalothorax, which is reddish brown with yellowish hairs in the species Tegenaria domestica (Figure 4). -

Biodiversity of the Huautla Cave System, Oaxaca, Mexico
diversity Communication Biodiversity of the Huautla Cave System, Oaxaca, Mexico Oscar F. Francke, Rodrigo Monjaraz-Ruedas † and Jesús A. Cruz-López *,‡ Colección Nacional De Arácnidos, Departamento de Zoología, Instituto de Biología, Universidad Nacional Autónoma de México, Ciudad Universitaria, Coyoacán, Mexico City C. P. 04510, Mexico; [email protected] (O.F.F.); [email protected] (R.M.-R.) * Correspondence: [email protected] † Current address: San Diego State University, San Diego, CA 92182, USA. ‡ Current address: Instituto Nacional de Investigaciones Agrícolas y Pecuarias del Valle de Oaxaca, Santo Domingo Barrio Bajo, Etla C. P. 68200, Mexico. Abstract: Sistema Huautla is the deepest cave system in the Americas at 1560 m and the fifth longest in Mexico at 89,000 m, and it is a mostly vertical network of interconnected passages. The surface landscape is rugged, ranging from 3500 to 2500 masl, intersected by streams and deep gorges. There are numerous dolinas, from hundreds to tens of meters in width and depth. The weather is basically temperate subhumid with summer rains. The average yearly rainfall is approximately 2500 mm, with a monthly average of 35 mm for the driest times of the year and up to 500 mm for the wettest month. All these conditions play an important role for achieving the highest terrestrial troglobite diversity in Mexico, containing a total of 35 species, of which 27 are possible troglobites (16 described), including numerous arachnids, millipedes, springtails, silverfish, and a single described species of beetles. With those numbers, Sistema Huautla is one of the richest cave systems in the world. Keywords: troglobitics; arachnids; insects; millipedes Citation: Francke, O.F.; Monjaraz-Ruedas, R.; Cruz-López, J.A. -

Salt and Water Balance in the Spider, Porrhothele Antipodiana (Mygalomorpha: Dipluridae): Effects of Feeding Upon Hydrated Animals
J. exp. Biol. 125, 85-106 (1986) 85 Printed in Great Britain © The Company of Biologists Limited 1986 SALT AND WATER BALANCE IN THE SPIDER, PORRHOTHELE ANTIPODIANA (MYGALOMORPHA: DIPLURIDAE): EFFECTS OF FEEDING UPON HYDRATED ANIMALS BY A. G. BUTT* AND H. H. TAYLORf Department of Zoology, University of Canterbury, Private Bag, Christchurch, New Zealand Accepted 15 May 1986 SUMMARY The spider, Porrhothele antipodiana, starved and provided with water, produced urine via the anal excretory system (Malpighian tubules, midgut diverticula and stercoral pocket) at a mean rate of about 2-5-5//I g"1 day"1 and with a mean Na+/K+ ratio of about 1-0. Salts ingested from the prey were eliminated by two mechanisms. A K+-rich (Na+/K+ about 0-2) anal diuresis lasted about 3 days following a single meal and was maintained at more than SO^lg^'day"1 during feeding ad libitum. The second mechanism, interpreted as coxal secretion, func- tioned only during feeding itself and delivered Na+ into the prey at a constant rate of about 3 %h~' of total body Na+. This progressively raised the Na+/K+ ratio of the prey debris from 0-47 to 0-96 and, because of re-ingestion, recycled more Na+ than was originally present in the prey. Feeding was associated with large net increases in dry weight and ions, particularly K+, which were mainly stored in the diverticular tissue (midgut diverticula and Malpighian tubules embedded in adipose tissue). The stercoral fluid (final urine) was slightly hyposmotic to the haemolymph in starved and fed spiders. Only about half of its osmolarity was accounted for by Na+, K+ and Cl~. -

The Complete Mitochondrial Genome of Endemic Giant Tarantula
www.nature.com/scientificreports OPEN The Complete Mitochondrial Genome of endemic giant tarantula, Lyrognathus crotalus (Araneae: Theraphosidae) and comparative analysis Vikas Kumar, Kaomud Tyagi *, Rajasree Chakraborty, Priya Prasad, Shantanu Kundu, Inderjeet Tyagi & Kailash Chandra The complete mitochondrial genome of Lyrognathus crotalus is sequenced, annotated and compared with other spider mitogenomes. It is 13,865 bp long and featured by 22 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs), 13 protein-coding genes (PCGs), and a control region (CR). Most of the PCGs used ATN start codon except cox3, and nad4 with TTG. Comparative studies indicated the use of TTG, TTA, TTT, GTG, CTG, CTA as start codons by few PCGs. Most of the tRNAs were truncated and do not fold into the typical cloverleaf structure. Further, the motif (CATATA) was detected in CR of nine species including L. crotalus. The gene arrangement of L. crotalus compared with ancestral arthropod showed the transposition of fve tRNAs and one tandem duplication random loss (TDRL) event. Five plesiomophic gene blocks (A-E) were identifed, of which, four (A, B, D, E) retained in all taxa except family Salticidae. However, block C was retained in Mygalomorphae and two families of Araneomorphae (Hypochilidae and Pholcidae). Out of 146 derived gene boundaries in all taxa, 15 synapomorphic gene boundaries were identifed. TreeREx analysis also revealed the transposition of trnI, which makes three derived boundaries and congruent with the result of the gene boundary mapping. Maximum likelihood and Bayesian inference showed similar topologies and congruent with morphology, and previously reported multi-gene phylogeny. However, the Gene-Order based phylogeny showed sister relationship of L. -
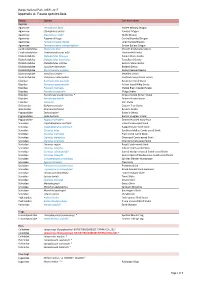
Report-Mungo National Park-Appendix A
Mungo National Park, NSW, 2017 Appendix A: Fauna species lists Family Species Common name Reptiles Agamidae Ctenophorus fordi Mallee Military Dragon Agamidae Ctenophorus pictus Painted Dragon Agamidae Diporiphora nobbi Nobbi Dragon Agamidae Pogona vitticeps Central Bearded Dragon Agamidae Tympanocryptis lineata Lined Earless Dragon Agamidae Tympanocryptis tetraporophora Eyrean Earless Dragon Carphodactylidae Nephrurus levis Smooth Knob-tailed Gecko Carphodactylidae Underwoodisaurus milii Thick-tailed Gecko Diplodactylidae Diplodactylus furcosus Ranges Stone Gecko Diplodactylidae Diplodactylus tessellatus Tessellated Gecko Diplodactylidae Diplodactylus vittatus Eastern Stone Gecko Diplodactylidae Lucasium damaeum Beaded Gecko Diplodactylidae Rhynchoedura ormsbyi Eastern Beaked Gecko Diplodactylidae Strophurus elderi ~ Jewelled Gecko Diplodactylidae Strophurus intermedius Southern Spiny-tailed Gecko Elapidae Brachyurophis australis Australian Coral Snake Elapidae Demansia psammophis Yellow-faced Whip Snake Elapidae Parasuta nigriceps Mallee Black-headed Snake Elapidae Pseudechis australis Mulga Snake Elapidae Pseudonaja aspidorhyncha * Strap-snouted Brown Snake Elapidae Pseudonaja textilis Eastern Brown Snake Elapidae Suta suta Curl Snake Gekkonidae Gehyra versicolor Eastern Tree Gecko Gekkonidae Heteronotia binoei Bynoe's Gecko Pygopodidae Delma butleri Butler's Delma Pygopodidae Lialis burtonis Burton's Legless Lizard Pygopodidae Pygopus schraderi Eastern Hooded Scaly-foot Scincidae Cryptoblepharus australis Inland Snake-eyed Skink Scincidae -

Tarantula Free Download
TARANTULA FREE DOWNLOAD Bob Dylan | 144 pages | 10 Jun 2011 | HarperCollins Publishers | 9780007215041 | English | London, United Kingdom Tarantulas They love warmer climates and that is why Tarantula are also in Mexico and portions of the United States. Entomology Expert. The name "tarantula" is also incorrectly applied to other large-bodied spiders, including Tarantula purseweb spiders or atypical tarantulasthe funnel-webs Dipluridae and Hexathelidaeand the " Tarantula tarantulas ". Tarantula big, beefy spiders strike fear in the hearts of arachnophobes everywhere, but in fact, tarantulas are some of the least Tarantula and Tarantula spiders around. Even though the Tarantula is usually a timid Spider, they have been known to show signs of aggression. Tarantula Deemer, who bluntly refuses permission. Also on the end of each leg, surrounding the claws, is a group of bristles, called the scopula, Tarantula help the tarantula to grip better when climbing surfaces such as glass. They weigh from 1 to 3 ounces Joe Burch Edwin Rand Hastings finds a Tarantula involving picked-clean cattle carcasses and large pools of a thick, white liquid. Last updated: February 24, Tarantula Director Jack Arnold used matte effects once again two years later to show miniaturization, rather than gigantism, in The Incredible Shrinking Manwhich also featured an encounter with a spider. Leonard Maltin's Classic Movie Guide. The Life of the Spider. The tarantula pursues them down the highway toward the town. Tarantula Eurypelma literally means 'wide footsole'; however, arachnologists have conventionally taken pelma in such names to refer to the scopula, so producing the meaning 'with a wide scopula'. These fine bristles are barbed Tarantula serve to irritate. -

(Araneae: Theraphosidae) from Miocene Chiapas Amber, Mexico
XX…………………………………… ARTÍCULO: A fossil tarantula (Araneae: Theraphosidae) from Miocene Chiapas amber, Mexico Jason A. Dunlop, Danilo Harms & David Penney ARTÍCULO: A fossil tarantula (Araneae: Theraphosidae) from Miocene Chiapas amber, Mexico Jason A. Dunlop Museum für Naturkunde der Humboldt Universität zu Berlin D-10115 Berlin, Germany [email protected] Abstract: Danilo Harms A fossil tarantula (Araneae: Mygalomorphae: Theraphosidae) is described from Freie Universität BerlinInstitut für an exuvium in Tertiary (Miocene) Chiapas amber, Simojovel region, Chiapas Biologie, Chemie & Pharmazie State, Mexico. It is difficult to assign it further taxonomically, but it is the first Evolution und Systematik der Tiere mygalomorph recorded from Chiapas amber and only the second unequivocal Königin-Luise-Str. 1–3 record of a fossil theraphosid. With a carapace length of ca. 0.9 cm and an es- D-14195 Berlin, Germany timated leg span of at least 5 cm it also represents the largest spider ever re- [email protected] corded from amber. Of the fifteen currently recognised mygalomorph families, eleven have a fossil record (summarised here), namely: Atypidae, Antrodiaeti- David Penney dae, Mecicobothriidae, Hexathelidae, Dipluridae, Ctenizidae, Nemesiidae, Mi- Earth, Atmospheric and Environmental crostigmatidae, Barychelidae, Cyrtaucheniidae and Theraphosidae. Sciences. Key words: Araneae, Theraphosidae, Palaeontology, Miocene, amber, Chiapas, The University of Manchester Mexico. Manchester. M13 9PL, UK [email protected] Revista Ibérica de Aracnología ISSN: 1576 - 9518. Un fósil de tarántula (Araneae: Theraphosidae) en ambar del Dep. Legal: Z-2656-2000. Vol. 15, 30-VI-2007 mioceno de Chiapas, México. Sección: Artículos y Notas. Pp: 9 − 17. Fecha publicación: 30 Abril 2008 Resumen: Se describe una tarántula fósil a partir de una exuvia en ámbar del terciario Edita: (mioceno) de Chiapas, región de Simojovel, estado de Chiapas, Mexico.