Transformation and Functional Verification of Cry5aa in Cotton
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

World Bank Document
Document of The World Bank FOR OFFICIAL USE ONLY Public Disclosure Authorized Report No: 42 124-CN PROJECT APPRAISAL DOCUMENT ON A PROPOSED LOAN Public Disclosure Authorized IN THE AMOUNT OF US$lOO MILLION TO THE PEOPLE’S REPUBLIC OF CHINA FOR A JIANGXI SHIHUTANG NAVIGATION AND HYDROPOWER COMPLEX PROJECT Public Disclosure Authorized August 14,2008 China and Mongolia Sustainable Development Unit Sustainable Development Department East Asia and Pacific Region This document has a restricted distribution and may be used by recipients only in the performance of their official duties. Its contents may not otherwise be disclosed without World Bank authorization. Public Disclosure Authorized CURRENCY EQUIVALENTS (Exchange Rate Effective January 28,2008) CurrencyUnit = RMB RMB1 = US$O.1389 US$1 = RMB7.2 FISCAL YEAR January 1 - December 31 ABBREVIATIONS AND ACRONYMS cccc China Communications Construction Company CDM Clean Development Mechanism CFU Carbon Finance Unit CGC China Guodian Corporation CPS Country Partnership Strategy CQS Selection Based on Consultants’ Qualifications DA Designated Account DP Displaced Person DSP Dam Safety Panel DWTIdwt Deadweight Tons EA Environmental Assessment EIA Environmental Impact Assessment EIRR Economic Internal Rate of Return EMP Environment Management Plan ENPV Economic Net Present Value EPP Emergency Preparedness Plan FM Financial Management FMS Financial Management Specialist FS Feasibility Study FSL Fixed Spread Loan FSR Feasibility Study Report FYP Five-Year Plan GDP Gross Domestic Product GOC Government -

Of the Chinese Bronze
READ ONLY/NO DOWNLOAD Ar chaeolo gy of the Archaeology of the Chinese Bronze Age is a synthesis of recent Chinese archaeological work on the second millennium BCE—the period Ch associated with China’s first dynasties and East Asia’s first “states.” With a inese focus on early China’s great metropolitan centers in the Central Plains Archaeology and their hinterlands, this work attempts to contextualize them within Br their wider zones of interaction from the Yangtze to the edge of the onze of the Chinese Bronze Age Mongolian steppe, and from the Yellow Sea to the Tibetan plateau and the Gansu corridor. Analyzing the complexity of early Chinese culture Ag From Erlitou to Anyang history, and the variety and development of its urban formations, e Roderick Campbell explores East Asia’s divergent developmental paths and re-examines its deep past to contribute to a more nuanced understanding of China’s Early Bronze Age. Campbell On the front cover: Zun in the shape of a water buffalo, Huadong Tomb 54 ( image courtesy of the Chinese Academy of Social Sciences, Institute for Archaeology). MONOGRAPH 79 COTSEN INSTITUTE OF ARCHAEOLOGY PRESS Roderick B. Campbell READ ONLY/NO DOWNLOAD Archaeology of the Chinese Bronze Age From Erlitou to Anyang Roderick B. Campbell READ ONLY/NO DOWNLOAD Cotsen Institute of Archaeology Press Monographs Contributions in Field Research and Current Issues in Archaeological Method and Theory Monograph 78 Monograph 77 Monograph 76 Visions of Tiwanaku Advances in Titicaca Basin The Dead Tell Tales Alexei Vranich and Charles Archaeology–2 María Cecilia Lozada and Stanish (eds.) Alexei Vranich and Abigail R. -
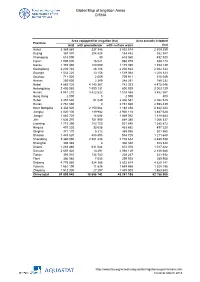
Global Map of Irrigation Areas CHINA
Global Map of Irrigation Areas CHINA Area equipped for irrigation (ha) Area actually irrigated Province total with groundwater with surface water (ha) Anhui 3 369 860 337 346 3 032 514 2 309 259 Beijing 367 870 204 428 163 442 352 387 Chongqing 618 090 30 618 060 432 520 Fujian 1 005 000 16 021 988 979 938 174 Gansu 1 355 480 180 090 1 175 390 1 153 139 Guangdong 2 230 740 28 106 2 202 634 2 042 344 Guangxi 1 532 220 13 156 1 519 064 1 208 323 Guizhou 711 920 2 009 709 911 515 049 Hainan 250 600 2 349 248 251 189 232 Hebei 4 885 720 4 143 367 742 353 4 475 046 Heilongjiang 2 400 060 1 599 131 800 929 2 003 129 Henan 4 941 210 3 422 622 1 518 588 3 862 567 Hong Kong 2 000 0 2 000 800 Hubei 2 457 630 51 049 2 406 581 2 082 525 Hunan 2 761 660 0 2 761 660 2 598 439 Inner Mongolia 3 332 520 2 150 064 1 182 456 2 842 223 Jiangsu 4 020 100 119 982 3 900 118 3 487 628 Jiangxi 1 883 720 14 688 1 869 032 1 818 684 Jilin 1 636 370 751 990 884 380 1 066 337 Liaoning 1 715 390 783 750 931 640 1 385 872 Ningxia 497 220 33 538 463 682 497 220 Qinghai 371 170 5 212 365 958 301 560 Shaanxi 1 443 620 488 895 954 725 1 211 648 Shandong 5 360 090 2 581 448 2 778 642 4 485 538 Shanghai 308 340 0 308 340 308 340 Shanxi 1 283 460 611 084 672 376 1 017 422 Sichuan 2 607 420 13 291 2 594 129 2 140 680 Tianjin 393 010 134 743 258 267 321 932 Tibet 306 980 7 055 299 925 289 908 Xinjiang 4 776 980 924 366 3 852 614 4 629 141 Yunnan 1 561 190 11 635 1 549 555 1 328 186 Zhejiang 1 512 300 27 297 1 485 003 1 463 653 China total 61 899 940 18 658 742 43 241 198 52 -

Bulliform Phytolith Research in Wild and Domesticated Rice Paddy Soil in South China
RESEARCH ARTICLE Bulliform Phytolith Research in Wild and Domesticated Rice Paddy Soil in South China Xiujia Huan1,2*, Houyuan Lu1,3*, Can Wang1,2, Xiangan Tang4, Xinxin Zuo1, Yong Ge1,2, Keyang He1,2 1 Institute of Geology and Geophysics, Chinese Academy of Sciences, Beijing, China, 2 University of Chinese Academy of Sciences, Beijing, China, 3 Center for Excellence in Tibetan Plateau Earth Science, Chinese Academy of Sciences, Beijing, China, 4 Soil & Fertilizer and Environmental & Resources Research Institute, Jiangxi Academy of Agricultural Sciences, Nanchang, Jiangxi Province, China * [email protected] (XH); [email protected] (HL) Abstract Bulliform phytoliths play an important role in researching rice origins as they can be used to distinguish between wild and domesticated rice. Rice bulliform phytoliths are character- OPEN ACCESS ized by numerous small shallow fish-scale decorations on the lateral side. Previous stud- Citation: Huan X, Lu H, Wang C, Tang X, Zuo X, Ge ies have shown that domesticated rice has a larger number of these decorations than wild Y, et al. (2015) Bulliform Phytolith Research in Wild rice and that the number of decorations 9 is a useful feature for identifying domesticated and Domesticated Rice Paddy Soil in South China. PLoS ONE 10(10): e0141255. doi:10.1371/journal. rice. However, this standard was established based on limited samples of modern rice pone.0141255 plants. In this study, we analyzed soil samples from both wild and domesticated rice pad- Editor: Xiaoyan Yang, Chinese Academy of dies. Results showed that, in wild rice soil samples, the proportion of bulliform phytoliths Sciences, CHINA with 9 decorations was 17.46% ± 8.29%, while in domesticated rice soil samples, the cor- ± Received: August 8, 2015 responding proportion was 63.70% 9.22%. -

Electrification Project of Zhe-Gan Railway Public Disclosure Authorized
! ~~~~~~~RP213Volume 2 Electrification Project of Zhe-Gan Railway Public Disclosure Authorized (Jiangxi Section) RESETTLEMENT ACTION PLAN Public Disclosure Authorized Public Disclosure Authorized Foreign-Financing Center of Ministry of Railways, PR China Public Disclosure Authorized East China Investigation and Design Institute I ~~~~under State Power Corporation May 2004 ;FILEFCwOPY Approved by: Gong Heping Examined by: Bian Bingqian Checked by: Yu Zhijian, Zhu Qiang Compiled by: Mao Zhenjun, Yu Zhijian, Zhu Qiang Xia Yunqiu, Gu Chunrong, Han Xiaojin, Li Min'an List of contents OBJECTIVES OF RESETJ'LEMENT PLAN & DEFINITION OF RESETTLEMENT TERMINOLOGY . 1 Generals ................................................................. 3 1.1 Project Background ...................................................... 3 1.1.1 Existing conditions ...................................................... 3 1.1.2 Necessity of of railway electrification ...................................................... 3 1.1.3 Significance of the project ...................................................... 5 1.2 Project Design and Design Approval .................................................... 6 1.3 Description of Project ...................................................... 7 1.4 Project Impacts ...................................................... 9 1.5 Measures To Alleviate Project Inpacts ....................................... 10 1.5.1 In project planning and design stage ..................................................... 10 1.5.2 In project implementation -

Chinese Pottery Traditions 19
CHINESE POTTERY TRADITIONS 19 Chinese Pottery Traditions Amoreena Rathke Faculty Sponsor: Karen Terpstra, Department of Art ABSTRACT My research was to conduct oral histories of Chinese potters in order to gain a bet- ter understanding of their role in the ceramics profession and their importance on my own artwork. The objective of my research of Chinese potters was multi-fold. Primarily, I was interested in anthropological data as it pertained to the present day to day life for Chinese potters, the role and function of ceramics in China, and especially the role that gender plays in the ceramic profession there. In addition, I was interested in the impact of traditional pottery techniques, forms, and glazes on Chinese potters and potters elsewhere in the world. I hoped to discover how those techniques are incorporated into modern ceramic work. Workspaces and studios were also an important research component as they are set up so differently than those in the United States. China has been quickly changing since the country opened up its doors to foreign travelers in 1976. Potters though out the world have greatly benefited by interactions with Chinese ceramicists. An unexpected result of the cultural exchange is that Chinese pottery is starting to break away from tradi- tion. I was greatly interested in researching modern potters as they begin a new ceramic dynasty in China. INTRODUCTION Ceramics have played an integral role in the lives of humans throughout history and con- tinue to enrich the human experience. China has a long and leading role in the evolution of ceramics and its cultural impact on the world. -

Document Produced Under TA: People's Republic of China: Jiangxi Rural Development Strategy Study
ADB Technical Assistance Project TA 7036-PRC Package 2: Jiangxi Development Strategy Jiangxi Rural Development Strategy Project Consulting Team Team leader: Wu Guobao © 2010 Asian Development Bank All rights reserved. Published 2010. Printed in the People's Republic of China The views expressed in this book are those of the authors and do not necessarily reflect the views and policies of the Asian Development Bank or its Board of Governors or the governments they represent. The Asian Development Bank does not guarantee the accuracy of the data included in this publication and accepts no responsibility for any consequence of their use. Use of the term “country” does not imply any judgment by the authors or the Asian Development Bank as to the legal or other status of any territorial entity. Contents Acknowledgements ....................................................................................v Contributors ...............................................................................................vi Abbreviations ........................................................................................... viii Executive Summary................................................................................... 1 I. Introduction ........................................................................................... 15 1. Context of the Study ........................................................................... 15 2. Objectives and Scope of the Study .................................................... 20 3. Approaches and -

Minimum Wage Standards in China August 11, 2020
Minimum Wage Standards in China August 11, 2020 Contents Heilongjiang ................................................................................................................................................. 3 Jilin ............................................................................................................................................................... 3 Liaoning ........................................................................................................................................................ 4 Inner Mongolia Autonomous Region ........................................................................................................... 7 Beijing......................................................................................................................................................... 10 Hebei ........................................................................................................................................................... 11 Henan .......................................................................................................................................................... 13 Shandong .................................................................................................................................................... 14 Shanxi ......................................................................................................................................................... 16 Shaanxi ...................................................................................................................................................... -

Negative Transfer of Zhangshu Dialect in Jiangxi Province to High School Students’ Acquisition of English Pronunciation
International Journal of Language and Linguistics Vol. 7, No. 4, December 2020 doi:10.30845/ijll.v7n4p7 Negative Transfer of Zhangshu Dialect in Jiangxi Province to High School Students’ Acquisition of English Pronunciation Wanlin Xu Faculty of Teacher Education Nanjing Normal University Nanjing, China Abstract Affected by the negative transfer of Zhangshu dialect in Jiangxi Province, China, many English learners are prone to errors in the acquisition of English pronunciation. In order to correct students’ wrong pronunciation and improve their pronunciation accuracy, this paper chose 30 senior high school students from Zhangshu Middle School to analyze their English pronunciation. The research results showed: (1) students were liable to confuse long monophthongs with the short ones and misread English vowels [æ] and [eə]; English consonants [θ], [ð], [tʃ] and [dʒ] were likely to be replaced by dialectal sounds [s], [ts], [tɕʰ] and [tɕ]respectively; English nasal [ŋ] in [iŋ] was also easy to be mispronounced. (2) At suprasegmental level, students tended to syllabicate the codas and stress each word in a sentence so that the rhythm featured dialectal characteristics. Also, their intonation was invariable due to the effect of Zhangshu dialect. Key words: Zhangshu dialect, senior high school students, negative transfer, English pronunciation Introduction Speaking ability, closely connected with reading, listening and writing abilities, is of great significance in the acquisition of a foreign language. As we all know, pronunciation is among the important factors to gauge students' speaking ability. Therefore, mastering its pronunciation will help students to achieve better outcomes in overall language acquisition. However, in the course of second language acquisition, regional dialects will exert negative effects on the articulation of the target language. -

Ceramic Tableware from China List of CNCA‐Certified Ceramicware
Ceramic Tableware from China June 15, 2018 List of CNCA‐Certified Ceramicware Factories, FDA Operational List No. 64 740 Firms Eligible for Consideration Under Terms of MOU Firm Name Address City Province Country Mail Code Previous Name XIAOMASHAN OF TAIHU MOUNTAINS, TONGZHA ANHUI HANSHAN MINSHENG PORCELAIN CO., LTD. TOWN HANSHAN COUNTY ANHUI CHINA 238153 ANHUI QINGHUAFANG FINE BONE PORCELAIN CO., LTD HANSHAN ECONOMIC DEVELOPMENT ZONE ANHUI CHINA 238100 HANSHAN CERAMIC CO., LTD., ANHUI PROVINCE NO.21, DONGXING STREET DONGGUAN TOWN HANSHAN COUNTY ANHUI CHINA 238151 WOYANG HUADU FINEPOTTERY CO., LTD FINEOPOTTERY INDUSTRIAL DISTRICT, SOUTH LIUQIAO, WOSHUANG RD WOYANG CITY ANHUI CHINA 233600 THE LISTED NAME OF THIS FACTORY HAS BEEN CHANGED FROM "SIU‐FUNG CERAMICS (CHONGQING SIU‐CERAMICS) CO., LTD." BASED ON NOTIFICATION FROM CNCA CHONGQING CHN&CHN CERAMICS CO., LTD. CHENJIAWAN, LIJIATUO, BANAN DISTRICT CHONGQING CHINA 400054 RECEIVED BY FDA ON FEBRUARY 8, 2002 CHONGQING KINGWAY CERAMICS CO., LTD. CHEN JIA WAN, LI JIA TUO, BANAN DISTRICT, CHONGQING CHINA 400054 BIDA CERAMICS CO.,LTD NO.69,CHENG TIAN SI GE DEHUA COUNTY FUJIAN CHINA 362500 NONE DATIAN COUNTY BAOFENG PORCELAIN PRODUCTS CO., LTD. YONGDE VILLAGE QITAO TOWN DATIAN COUNTY CHINA 366108 FUJIAN CHINA DATIAN YONGDA ART&CRAFT PRODUCTS CO., LTD. NO.156, XIANGSHAN ROAD, JUNXI TOWN, DATIAN COUNTY FUJIAN 366100 DEHUA KAIYUAN PORCELAIN INDUSTRY CO., LTD NO. 63, DONGHUAN ROAD DEHUA TOWN FUJIAN CHINA 362500 THE LISTED ADDRESS OF THIS FACTORY HAS BEEN CHANGED FROM "MAQIUYANG XUNZHONG XUNZHONG TOWN, DEHUA COUNTY" TO THE NEW EAST SIDE, THE SECOND PERIOD, SHIDUN PROJECT ADDRESS LISTED ABOVE BASED ON NOTIFICATION DEHUA HENGHAN ARTS CO., LTD AREA, XUNZHONG TOWN, DEHUA COUNTY FUJIAN CHINA 362500 FROM THE CNCA AUTHORITY IN SEPTEMBER 2014 DEHUA HONGSHENG CERAMICS CO., LTD. -

CHINESE CERAMICS and TRADE in 14 CENTURY SOUTHEAST ASIA——A CASE STUDY of SINGAPORE XIN GUANGCAN (BA History, Pekingu;MA Arch
CHINESE CERAMICS AND TRADE IN 14TH CENTURY SOUTHEAST ASIA——A CASE STUDY OF SINGAPORE XIN GUANGCAN (BA History, PekingU;MA Archaeology, PekingU) A THESIS SUBMITTED FOR THE DEGREE OF DOCTOR OF PHILOSOPHY DEPARTMENT OF SOUTHEAST ASIAN STUDIES NATIONAL UNIVERSITY OF SINGAPORE 2015 Acknowledgements Upon accomplishing the entire work of this thesis, it is time for me to acknowledge many people who have helped me. First, I will like to express my utmost gratitude to my supervisor Dr.John N. Miksic from the Department of Southeast Asian Studies, National University of Singapore. He has dedicated a lot of precious time to supervising me, from choosing the thesis topic, organizing the fieldwork plans, to giving much valued comments and advice on the immature thesis drafts. I am the most indebted to him. The committee member Dr. Patric Daly from the Asian Research Institute and Dr. Yang Bin from the History Department, who gave me useful suggestions during the qualifying examination. I also would like to thank the following people who have given me a lot of support during my fieldwork and final stage of writing. For the fieldtrip in Zhejiang Province, with the help of Mr. Shen Yueming, the director of Zhejiang Relics and Archaeology Institute, I was able to be involved in a meaningful excavation of a Song to Yuan Dynasty ceramic kiln site in Longquan County. During the excavation, the deputy team leader Mr. Xu Jun and the local researcher Mr. Zhou Guanggui gave a lot of suggestions on the identification of Longquan celadon. Moreover, Ms. Wu Qiuhua, Mr. Yang Guanfu, and Mr. -

Big Ding 鼎 and China Power: Divine Authority and Legitimacy
Big Ding 鼎 and China Power: Divine Authority and Legitimacy ELIZABETH CHILDS-JOHNSON By the eastern zhou and imperial eras of Chinese history, a legend had grown cel- ebrating the ding 鼎 bronze vessel as the preeminent symbol of state authority and divine power. The mythic theme of “The First Emperor’s [Qin Shi Huangdi’s] Search for the Zhou Ding” or “The First Qin Emperor’s Failure to Discover the Ding” deco- rate the main gables of more than several Eastern Han funerary shrines, including Xiaotangshan and Wuliang in Shandong province (Wu 1989 : 138, 348). Pre-Han records in the Zuozhuan: 7th year of Duke Zhao (左传: 昭公七年) as well as the “Geng- zhu” chapter in the Mozi (墨子: 耕柱篇) record the significance of this mythic representation. The Mozi passage states: In ancient times, King Qi of the Xia [Xia Qi Wang] commissioned Feilian to dig minerals in mountains and rivers and to use clay molds, casting the ding at Kunwu. He ordered Wengnanyi to divine with the help of the tortoise from Bairuo, saying: “Let the ding, when completed, have a square body and four legs. Let them be able to boil without kindling, to hide themselves without being lifted, and to move themselves without being carried so that they will be used for sacrifice at Kunwu.” Yi interpreted the oracle as saying: “The offering has been accepted. When the nine ding have been completed, they will be ‘transferred’ down to three kingdoms. When Xia loses them, people of the Yin will possess them, and when people of the Yin lose them, people of the Zhou will pos- sess them.”1 [italics added] As maintained in this article, the inspiration for this popular legend of mythic power most likely originated during dynastic Shang times with the first casting in bronze of the monumental, four-legged ding.