Gene Pool of Buryats: Clinal Variability and Territorial Subdivision Based on Data of Y�Chromosome Markers V
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
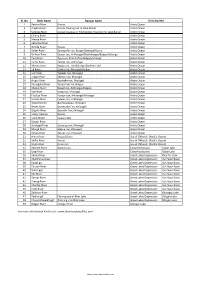
List of Rivers of Mongolia
Sl. No River Name Russian Name Draining Into 1 Yenisei River Russia Arctic Ocean 2 Angara River Russia, flowing out of Lake Baikal Arctic Ocean 3 Selenge River Сэлэнгэ мөрөн in Sükhbaatar, flowing into Lake Baikal Arctic Ocean 4 Chikoy River Arctic Ocean 5 Menza River Arctic Ocean 6 Katantsa River Arctic Ocean 7 Dzhida River Russia Arctic Ocean 8 Zelter River Зэлтэрийн гол, Bulgan/Selenge/Russia Arctic Ocean 9 Orkhon River Орхон гол, Arkhangai/Övörkhangai/Bulgan/Selenge Arctic Ocean 10 Tuul River Туул гол, Khentii/Töv/Bulgan/Selenge Arctic Ocean 11 Tamir River Тамир гол, Arkhangai Arctic Ocean 12 Kharaa River Хараа гол, Töv/Selenge/Darkhan-Uul Arctic Ocean 13 Eg River Эгийн гол, Khövsgöl/Bulgan Arctic Ocean 14 Üür River Үүрийн гол, Khövsgöl Arctic Ocean 15 Uilgan River Уйлган гол, Khövsgöl Arctic Ocean 16 Arigiin River Аригийн гол, Khövsgöl Arctic Ocean 17 Tarvagatai River Тарвагтай гол, Bulgan Arctic Ocean 18 Khanui River Хануй гол, Arkhangai/Bulgan Arctic Ocean 19 Ider River Идэр гол, Khövsgöl Arctic Ocean 20 Chuluut River Чулуут гол, Arkhangai/Khövsgöl Arctic Ocean 21 Suman River Суман гол, Arkhangai Arctic Ocean 22 Delgermörön Дэлгэрмөрөн, Khövsgöl Arctic Ocean 23 Beltes River Бэлтэсийн Гол, Khövsgöl Arctic Ocean 24 Bügsiin River Бүгсийн Гол, Khövsgöl Arctic Ocean 25 Lesser Yenisei Russia Arctic Ocean 26 Kyzyl-Khem Кызыл-Хем Arctic Ocean 27 Büsein River Arctic Ocean 28 Shishged River Шишгэд гол, Khövsgöl Arctic Ocean 29 Sharga River Шарга гол, Khövsgöl Arctic Ocean 30 Tengis River Тэнгис гол, Khövsgöl Arctic Ocean 31 Amur River Russia/China -

Germanic Origins from the Perspective of the Y-Chromosome
Germanic Origins from the Perspective of the Y-Chromosome By Michael Robert St. Clair A dissertation submitted in partial satisfaction of the requirements for the degree of Doctor in Philosophy in German in the Graduate Division of the University of California, Berkeley Committee in charge: Irmengard Rauch, Chair Thomas F. Shannon Montgomery Slatkin Spring 2012 Abstract Germanic Origins from the Perspective of the Y-Chromosome by Michael Robert St. Clair Doctor of Philosophy in German University of California, Berkeley Irmengard Rauch, Chair This dissertation holds that genetic data are a useful tool for evaluating contemporary models of Germanic origins. The Germanic languages are a branch of the Indo-European language family and include among their major contemporary representatives English, German, Dutch, Danish, Swedish, Norwegian and Icelandic. Historically, the search for Germanic origins has sought to determine where the Germanic languages evolved, and why the Germanic languages are similar to and different from other European languages. Both archaeological and linguist approaches have been employed in this research direction. The linguistic approach to Germanic origins is split among those who favor the Stammbaum theory and those favoring language contact theory. Stammbaum theory posits that Proto-Germanic separated from an ancestral Indo-European parent language. This theoretical approach accounts for similarities between Germanic and other Indo- European languages by posting a period of mutual development. Germanic innovations, on the other hand, occurred in isolation after separation from the parent language. Language contact theory posits that Proto-Germanic was the product of language convergence and this convergence explains features that Germanic shares with other Indo-European languages. -

Mitochondrial Haplogroup Background May Influence
Genetics Mitochondrial Haplogroup Background May Influence Southeast Asian G11778A Leber Hereditary Optic Neuropathy Supannee Kaewsutthi,1,2 Nopasak Phasukkijwatana,2,3 Yutthana Joyjinda,1 Wanicha Chuenkongkaew,3,4 Bussaraporn Kunhapan,1 Aung Win Tun,1 Bhoom Suktitipat,1 and Patcharee Lertrit1,4 PURPOSE. To investigate the role of mitochondrial DNA markedly incomplete penetrance. The three most common (mt DNA) background on the expression of Leber hereditary primary LHON mutations, G3460A in ND1, G11778A in ND4, optic neuropathy (LHON) in Southeast Asian carriers of the and T14484C in ND6, account for more than 90% of LHON G11778A mutation. cases worldwide2 with G11778A being the most common. In 3 4–6 METHODS. Complete mtDNA sequences were analyzed from 53 Thailand and other Asian countries, G11778A is responsi- unrelated Southeast Asian G11778A LHON pedigrees in Thai- ble for approximately 90% of LHON families. land and 105 normal Thai controls, and mtDNA haplogroups The sex bias and the marked incomplete penetrance of were determined. Clinical phenotypes were tested for associ- LHON indicate that there must be other factors that modify disease expression. Mitochondrial background,7–8 nuclear ation with mtDNA haplogroup, with adjustment for potential 9–11 12 confounders such as sex and age at onset. background, and environmental factors have been impli- cated in disease expression, although the precise mechanisms RESULTS. mtDNA subhaplogroup B was significantly associated of pathogenesis are largely undefined. with LHON. Follow-up analysis -

Preserving the Symbol of Siberia, Moving On: Sobol' and The
EA-13 • RUSSIA • JULY 2009 ICWA Letters INSTITUTE OF CURRENT WORLD AFFAIRS Preserving the Symbol of Siberia, Moving On: Sobol’ and the Elena Agarkova is studying management Barguzinsky Zapovednik (Part I) of natural resources and the relationship between By Elena Agarkova Siberia’s natural riches and its people. Previously, Elena was a Legal Fellow at the LAKE BAIKAL–I started researching this news- University of Washington’s letter with a plan to write about the Barguzin- School of Law, at the sky zapovednik, a strict nature reserve on the Berman Environmental eastern shore of Baikal, the first and the old- Law Clinic. She has clerked est in the country.1 I went to Nizhneangarsk, a for Honorable Cynthia M. Rufe of the federal district small township at the north shore of the lake, court in Philadelphia, and where the zapovednik’s head office is located has practiced commercial now. I crossed the lake and hiked on the east- litigation at the New York ern side through some of the zapovednik’s ter- office of Milbank, Tweed, ritory. I talked to people who devoted their lives Hadley & McCloy LLP. Elena to preserving a truly untouched wilderness, on was born in Moscow, Rus- a shoestring budget. And along the way I found sia, and has volunteered for myself going in a slightly different direction environmental non-profits than originally planned. An additional protago- in the Lake Baikal region of Siberia. She graduated nist emerged. I became fascinated by a small, from Georgetown Universi- elusive animal that played a central role not ty Law Center in 2001, and only in the creation of Russia’s first strict nature has received a bachelor’s reserve, but in the history of Russia itself. -

Narratives of Social Change in Rural Buryatia, Russia
Georgia State University ScholarWorks @ Georgia State University Anthropology Theses Department of Anthropology 4-20-2010 Narratives of Social Change in Rural Buryatia, Russia Luis Ortiz-Echevarria Georgia State University Follow this and additional works at: https://scholarworks.gsu.edu/anthro_theses Part of the Anthropology Commons Recommended Citation Ortiz-Echevarria, Luis, "Narratives of Social Change in Rural Buryatia, Russia." Thesis, Georgia State University, 2010. https://scholarworks.gsu.edu/anthro_theses/36 This Thesis is brought to you for free and open access by the Department of Anthropology at ScholarWorks @ Georgia State University. It has been accepted for inclusion in Anthropology Theses by an authorized administrator of ScholarWorks @ Georgia State University. For more information, please contact [email protected]. NARRATIVES OF SOCIAL CHANGE IN RURAL BURYATIA, RUSSIA by LUIS RAIMUNDO JESÚS ORTIZ ECHEVARRÍA Under the Direction of Jennifer Patico ABSTRACT This study explores postsocialist representations of modernity and identity through narratives of social change collected from individuals in rural communities of Buryatia, Russia. I begin with an examination of local conceptualizations of the past, present, and future and how they are imagined in places and spaces. Drawing on 65 days of fieldwork, in-depth interviews, informal discussion, and participant-observation, I elaborate on what I am calling a confrontation with physical triggers of self in connection to place, including imaginations of the countryside and -

CORE STRENGTH WITHIN MONGOL DIASPORA COMMUNITIES Archaeological Evidence Places Early Stone Age Human Habitation in the Southern
CORE STRENGTH WITHIN MONGOL DIASPORA COMMUNITIES Archaeological evidence places early Stone Age human habitation in the southern Gobi between 100,000 and 200,000 years ago 1. While they were nomadic hunter-gatherers it is believed that they migrated to southern Asia, Australia, and America through Beringia 50,000 BP. This prehistoric migration played a major role in fundamental dispersion of world population. As human migration was an essential part of human evolution in prehistoric era the historical mass dispersions in Middle Age and Modern times brought a significant influence on political and socioeconomic progress throughout the world and the latter has been studied under the Theory of Diaspora. This article attempts to analyze Mongol Diaspora and its characteristics. The Middle Age-Mongol Diaspora started by the time of the Great Mongol Empire was expanding from present-day Poland in the west to Korea in the east and from Siberia in the north to the Gulf of Oman and Vietnam in the south. Mongols were scattered throughout the territory of the Great Empire, but the disproportionately small number of Mongol conquerors compared with the masses of subject peoples and the change in Mongol cultural patterns along with influence of foreign religions caused them to fell prey to alien cultures after the decline of the Empire. As a result, modern days Hazara communities in northeastern Afghanistan and a small group of Mohol/Mohgul in India, Daur, Dongxiang (Santa), Monguor or Chagaan Monggol, Yunnan Mongols, Sichuan Mongols, Sogwo Arig, Yugur and Bonan people in China are considered as descendants of Mongol soldiers, who obeyed their Khaan’s order to safeguard the conquered area and waited in exceptional loyalty. -

An Overview of the Independent Histories of the Human Y Chromosome and the Human Mitochondrial Chromosome
The Proceedings of the International Conference on Creationism Volume 8 Print Reference: Pages 133-151 Article 7 2018 An Overview of the Independent Histories of the Human Y Chromosome and the Human Mitochondrial chromosome Robert W. Carter Stephen Lee University of Idaho John C. Sanford Cornell University, Cornell University College of Agriculture and Life Sciences School of Integrative Plant Science,Follow this Plant and Biology additional Section works at: https://digitalcommons.cedarville.edu/icc_proceedings DigitalCommons@Cedarville provides a publication platform for fully open access journals, which means that all articles are available on the Internet to all users immediately upon publication. However, the opinions and sentiments expressed by the authors of articles published in our journals do not necessarily indicate the endorsement or reflect the views of DigitalCommons@Cedarville, the Centennial Library, or Cedarville University and its employees. The authors are solely responsible for the content of their work. Please address questions to [email protected]. Browse the contents of this volume of The Proceedings of the International Conference on Creationism. Recommended Citation Carter, R.W., S.S. Lee, and J.C. Sanford. An overview of the independent histories of the human Y- chromosome and the human mitochondrial chromosome. 2018. In Proceedings of the Eighth International Conference on Creationism, ed. J.H. Whitmore, pp. 133–151. Pittsburgh, Pennsylvania: Creation Science Fellowship. Carter, R.W., S.S. Lee, and J.C. Sanford. An overview of the independent histories of the human Y-chromosome and the human mitochondrial chromosome. 2018. In Proceedings of the Eighth International Conference on Creationism, ed. J.H. -

N.I.Il`Minskii and the Christianization of the Chuvash
Durham E-Theses Narodnost` and Obshchechelovechnost` in 19th century Russian missionary work: N.I.Il`minskii and the Christianization of the Chuvash KOLOSOVA, ALISON,RUTH How to cite: KOLOSOVA, ALISON,RUTH (2016) Narodnost` and Obshchechelovechnost` in 19th century Russian missionary work: N.I.Il`minskii and the Christianization of the Chuvash, Durham theses, Durham University. Available at Durham E-Theses Online: http://etheses.dur.ac.uk/11403/ Use policy The full-text may be used and/or reproduced, and given to third parties in any format or medium, without prior permission or charge, for personal research or study, educational, or not-for-prot purposes provided that: • a full bibliographic reference is made to the original source • a link is made to the metadata record in Durham E-Theses • the full-text is not changed in any way The full-text must not be sold in any format or medium without the formal permission of the copyright holders. Please consult the full Durham E-Theses policy for further details. Academic Support Oce, Durham University, University Oce, Old Elvet, Durham DH1 3HP e-mail: [email protected] Tel: +44 0191 334 6107 http://etheses.dur.ac.uk 2 1 Narodnost` and Obshchechelovechnost` in 19th century Russian missionary work: N.I.Il`minskii and the Christianization of the Chuvash PhD Thesis submitted by Alison Ruth Kolosova Material Abstract Nikolai Il`minskii, a specialist in Arabic and the Turkic languages which he taught at the Kazan Theological Academy and Kazan University from the 1840s to 1860s, became in 1872 the Director of the Kazan Teachers‟ Seminary where the first teachers were trained for native- language schools among the Turkic and Finnic peoples of the Volga-Urals and Siberia. -

Ancient Mitochondrial DNA from Pre-Historic
Grand Valley State University ScholarWorks@GVSU Masters Theses Graduate Research and Creative Practice 4-30-2011 Ancient Mitochondrial DNA From Pre-historic Southeastern Europe: The rP esence of East Eurasian Haplogroups Provides Evidence of Interactions with South Siberians Across the Central Asian Steppe Belt Jeremy R. Newton Grand Valley State University Follow this and additional works at: http://scholarworks.gvsu.edu/theses Part of the Cell Biology Commons, and the Molecular Biology Commons Recommended Citation Newton, Jeremy R., "Ancient Mitochondrial DNA From Pre-historic Southeastern Europe: The rP esence of East Eurasian Haplogroups Provides Evidence of Interactions with South Siberians Across the Central Asian Steppe Belt" (2011). Masters Theses. 5. http://scholarworks.gvsu.edu/theses/5 This Thesis is brought to you for free and open access by the Graduate Research and Creative Practice at ScholarWorks@GVSU. It has been accepted for inclusion in Masters Theses by an authorized administrator of ScholarWorks@GVSU. For more information, please contact [email protected]. ANCIENT MITOCHONDRIAL DNA FROM PRE-HISTORIC SOUTH- EASTERN EUROPE: THE PRESENCE OF EAST EURASIAN HAPLOGROUPS PROVIDES EVIDENCE OF INTERACTIONS WITH SOUTH SIBERIANS ACROSS THE CENTRAL ASIAN STEPPE BELT A thesis submittal in partial fulfillment of the requirements for the degree of Master of Science By Jeremy R. Newton To Cell and Molecular Biology Department Grand Valley State University Allendale, MI April, 2011 “Not all those who wander are lost.” J.R.R. Tolkien iii ACKNOWLEDGEMENTS I would like to extend my sincerest thanks to every person who has motivated, directed, and encouraged me throughout this thesis project. I especially thank my graduate advisor, Dr. -

The Petroleum Potential of the Riphean–Vendian Succession of Southern East Siberia
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/253369249 The petroleum potential of the Riphean–Vendian succession of southern East Siberia CHAPTER in GEOLOGICAL SOCIETY LONDON SPECIAL PUBLICATIONS · MAY 2012 Impact Factor: 2.58 · DOI: 10.1144/SP366.1 CITATIONS READS 2 95 4 AUTHORS, INCLUDING: Olga K. Bogolepova Uppsala University 51 PUBLICATIONS 271 CITATIONS SEE PROFILE Alexander P. Gubanov Scandiz Research 55 PUBLICATIONS 485 CITATIONS SEE PROFILE Available from: Olga K. Bogolepova Retrieved on: 08 March 2016 Downloaded from http://sp.lyellcollection.org/ by guest on March 25, 2013 Geological Society, London, Special Publications The petroleum potential of the Riphean-Vendian succession of southern East Siberia James P. Howard, Olga K. Bogolepova, Alexander P. Gubanov and Marcela G?mez-Pérez Geological Society, London, Special Publications 2012, v.366; p177-198. doi: 10.1144/SP366.1 Email alerting click here to receive free e-mail alerts when service new articles cite this article Permission click here to seek permission to re-use all or request part of this article Subscribe click here to subscribe to Geological Society, London, Special Publications or the Lyell Collection Notes © The Geological Society of London 2013 Downloaded from http://sp.lyellcollection.org/ by guest on March 25, 2013 The petroleum potential of the Riphean–Vendian succession of southern East Siberia JAMES P. HOWARD*, OLGA K. BOGOLEPOVA, ALEXANDER P. GUBANOV & MARCELA GO´ MEZ-PE´ REZ CASP, West Building, 181a Huntingdon Road, Cambridge CB3 0DH, UK *Corresponding author (e-mail: [email protected]) Abstract: The Siberian Platform covers an area of c. -

RCN #33 21/8/03 13:57 Page 1
RCN #33 21/8/03 13:57 Page 1 No. 33 Summer 2003 Special issue: The Transformation of Protected Areas in Russia A Ten-Year Review PROMOTING BIODIVERSITY CONSERVATION IN RUSSIA AND THROUGHOUT NORTHERN EURASIA RCN #33 21/8/03 13:57 Page 2 CONTENTS CONTENTS Voice from the Wild (Letter from the Editors)......................................1 Ten Years of Teaching and Learning in Bolshaya Kokshaga Zapovednik ...............................................................24 BY WAY OF AN INTRODUCTION The Formation of Regional Associations A Brief History of Modern Russian Nature Reserves..........................2 of Protected Areas........................................................................................................27 A Glossary of Russian Protected Areas...........................................................3 The Growth of Regional Nature Protection: A Case Study from the Orlovskaya Oblast ..............................................29 THE PAST TEN YEARS: Making Friends beyond Boundaries.............................................................30 TRENDS AND CASE STUDIES A Spotlight on Kerzhensky Zapovednik...................................................32 Geographic Development ........................................................................................5 Ecotourism in Protected Areas: Problems and Possibilities......34 Legal Developments in Nature Protection.................................................7 A LOOK TO THE FUTURE Financing Zapovedniks ...........................................................................................10 -

Russian-Chinese Trade in Kyakhta — Trade Development and Volume Indicators 1727–1861
Russian-Chinese Trade in Kyakhta — Trade Development and Volume Indicators 1727–1861 Michal Wanner This study is a follow-up to the author’s previous articles featuring the beginning of the Russian-Chinese diplomatic and trade relations, and the early forms of the Rus- sian-Chinese trade,1 as well as characteristic of the Russian-Chinese trade exchange organisation and practice in Kyakhta and May-ma-chen, or both Russian and Chi- OPEN ACCESS nese export commodity structure; but also to a few fundamental logistics issues hav- ing considerable impact on character of the trade in Kyakhta.2 The author focused this, third in row but last study, on portions of this broad topic, which had not been possible to include in the previous articles for the reasons of their size, specifically, the trade quantification and capturing single stages development in the monitored period, the trade influence on the overall advance of the region, and also the reasons that resulted in the decline and end of the Russian-Chinese trade in Kyakhta, more specifically, on other forms of the trade exchange as the replacement. THE TRADE DEVELOPMENT AND VOLUME In December 1728, shortly after The Treaty of Kyakhta had been entered into, Chinese merchants began to have complaints about the shortage of Russian merchants and their goods, but also because they had been only selling cloth and Russian leather. However, the situation would shortly change. Russian goods on 1,430 carriages and 96 sleighs were delivered to Kyakhta between 1736 and 1740, whereas Chinese goods were carried on 806 carriages and 37 sleighs.3 1 M.