Application of Artificial Intelligence Technology in Oncology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Effective Application of Digital Printing Techniques for Fine Artists in the South African Context
The effective application of digital printing techniques for fine artists in the South African context. a Dissertation BY Susan Louise Giloi (NH Dip: Photography) Submitted in partial compliance with the requirements for the Degree Magister Technologiae in Photography in the Department of Photography, Faculty of Art and Design. PORT ELIZABETH TECHNIKON November 1999 PROMOTERS Prof. N.P.L. Allen M.F.A. D.Phil. Laur Tech Mr S.C. Strooh N Dip: Photography DEDICATION To my family who gave me the opportunity to do this. To Prof. Nicholas Allen and Mr. Steven Strooh for their help and inspiration. ii ACKNOWLEDGEMENTS My thanks to all the individuals and companies that helped me. Ashley Bowers, Tone Graphics Billy Whitehair, Printing Products Bonny Lhotka Bruce Cadle, Port Elizabeth Technikon Carol Van Zyl, Vaal Triangle Technikon Cleone Cull, Port Elizabeth Technikon Colleen Bate, Digital Imaging and Publishing Dean Johnstone, Square One Dianna Wall, Museum Africa Dorothy Krause Edwin Taylor, Beith Digital Eileen Fritsch, Big Picture Magazine Ethna Frankenfeld, Port Elizabeth Technikon Eugene Pienaar, Port Elizabeth Technikon Gavin Van Rensburg, Kemtek Geoff Black, Cyber Distributors Glen Meyer, Port Elizabeth Technikon Hank Van Der Water, Xerox E.C. Ian de Vega, Port Elizabeth Technikon Ian Marley, Vaal Triangle Technikon Ian White, John Cook University Inge Economou, Port Elizabeth Technikon Isabel Lubbe, Vaal Triangle Technikon Isabel Smit, Yeltech Jenny Ord, Port Elizabeth Technikon John Clarke, JFC Clarke Studio John Otsuki, Capitol Color Keith Solomon, First Graphics Laraine Bekker, Port Elizabteh Technikon Lisle Nel, Port Elizabeth Technikon Magda Bosch, Port Elizabteh Technikon Mary Duker, Port Elizabeth Technikon Mike Swanepoel, Port Elizabeth Technikon Nick Hand, Nexus Nolan Weight, Stonehouse Graphics Peter Thome, Agfa SA Robin Mowatt, QMS Ronald Henry, Omni Graphics Ros Streak, City Graphics Russell Williams, Teltron Steve Majewski Thinus Mathee, Vaal Triangle Technikon iii Tilla Jordaan, Xerox SA Tony Davidson, Outdoor Advertising Association of S.A. -
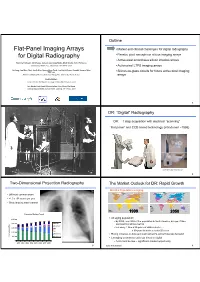
Flat-Panel Imaging Arrays for Digital Radiography
Outline Flat-Panel Imaging Arrays • Market and clinical challenges for digital radiography for Digital Radiography • Passive pixel amorphous silicon imaging arrays • Active pixel amorphous silicon imaging arrays Timothy Tredwell, Jeff Chang, Jackson Lai, Greg Heiler, Mark Shafer, John Yorkston Carestream Health, Inc., Rochester, NY 14615, USA • Active pixel LTPS imaging arrays Jin Jang, Jae Won Choi, Jae Ik Kim, Seung Hyun Park, Jun Hyuk Cheon, Sauabh Saxena, Won Kyu Lee • Silicon-on-glass circuits for future active pixel imaging Advanced Display Research Center, Kyung Hee University, Seoul, Korea arrays Arokia Nathan London Center for Nanotechnology, University College, London Eric Mozdy, Carlo Kosik Williams, Jeffery Cites, Chuan Che Wang Corning Incorporated, Sullivan Park, Corning, NY 14831, USA 2 DR: “Digital” Radiography DR: 1 step acquisition with electrical “scanning” “Flat panel” and CCD based technology (introduced ~1995) (Courtesy Imaging Dynamics Corp.) 3 4 Two-Dimensional Projection Radiography The Market Outlook for DR: Rapid Growth World’s Population is Aging • Still most common exam • >1.5 x 10 9 exams per year • Chest imaging most common 1999 2050 Procedural Volume Trends • An aging population: 2,500 • By 2050, over 25% of the population in North America, Europe, China 2,000 Nuc Med and Australia will be over 60 ULtrasound • For every 1 time a 20-year-old visits a doctor … 1,500 MR …a 60-year-old visits a doctor 26 times 1,000 CT Digital X-ray • Rising incomes in Asia and Latin America will accelerate demand Procedures (Ms) Procedures 500 Analog x-ray • Emerging economies could go direct to digital - • Cost must be low – significant market opportunity 2001 2002 2003 2004 2005 2006 2007 2008 5 Source: WHO, World Bank 6 Anatomical Noise Anatomical Noise in Projection Radiography 3-Dim 2-Dim &KHVW5DGLRJUDSK 0DPPRJUDSK\ • 3 dim. -

Prominence of Expert System and Case Study- DENDRAL Namita Mirjankar, Shruti Ghatnatti Karnataka, India [email protected],[email protected]
International Journal of Advanced Networking & Applications (IJANA) ISSN: 0975-0282 Prominence of Expert System and Case Study- DENDRAL Namita Mirjankar, Shruti Ghatnatti Karnataka, India [email protected],[email protected] Abstract—Among many applications of Artificial Intelligence, Expert System is the one that exploits human knowledge to solve problems which ordinarily would require human insight. Expert systems are designed to carry the insight and knowledge found in the experts in a particular field and take decisions based on the accumulated knowledge of the knowledge base along with an arrangement of standards and principles of inference engine, and at the same time, justify those decisions with the help of explanation facility. Inference engine is continuously updated as new conclusions are drawn from each new certainty in the knowledge base which triggers extra guidelines, heuristics and rules in the inference engine. This paper explains the basic architecture of Expert System , its first ever success DENDRAL which became a stepping stone in the Artificial Intelligence field, as well as the difficulties faced by the Expert Systems Keywords—Artificial Intelligence; Expert System architecture; knowledge base; inference engine; DENDRAL I INTRODUCTION the main components that remain same are: User Interface, Knowledge Base and Inference Engine. For more than two thousand years, rationalists all over the world have been striving to comprehend and resolve two unavoidable issues of the universe: how does a human mind work, and can non-people have minds? In any case, these inquiries are still unanswered. As humans, we all are blessed with the ability to learn and comprehend, to think about different issues and to decide; but can we design machines to do all these things? Some philosophers are open to the idea that machines will perform all the tasks a human can do. -

Digital and Advanced Imaging Equipment
CHAPTER 9 Digital and Advanced Imaging Equipment KEY TERMS active matrix array direct-to-digital radiographic systems photostimulated luminescence amorphous dual-energy x-ray absorptiometry picture archiving and communication analog-to-digital converter F-center system aspect ratio fill factor preprocessing cinefluorography frame rate postprocessing computed radiography image contrast refresh rate detective quantum efficiency image enhancement special procedures laboratory Digital Imaging and Communications image management and specular reflection in Medicine group communication system teleradiology digital fluoroscopy image restoration thin-film transistor digital radiography interpolation window level digital subtraction angiography liquid crystal display window width digital x-ray radiogrammetry Nyquist frequency OBJECTIVES At the completion of this chapter the reader should be able to do the following: • Describe the basic methods of obtaining digital cathode-ray tube cameras, videotape and videodisc radiographs recorders, and cinefluorographic equipment and discuss • State the advantages and disadvantages of digital the quality control procedures for each radiography versus conventional film/screen • Describe the various types of electronic display devices radiography and discuss the applicable quality control procedures • Discuss the quality control procedures for evaluating • Explain the basic image archiving and management digital radiographic systems networks and discuss the applicable quality control • Describe the basic methods -

Overview of Digital Detector Technology
Overview of Digital Detector Technology J. Anthony Seibert, Ph.D. Department of Radiology University of California, Davis Disclosure • Member (uncompensated) – Barco-Voxar Medical Advisory Board – ALARA (CR manufacturer) Advisory Board Learning Objectives • Describe digital versus screen-film acquisition • Introduce digital detector technologies • Compare cassette and cassette-less operation in terms of resolution, efficiency, noise • Describe new acquisition & processing techniques • Discuss PACS/RIS interfaces and features 1 Conventional screen/film detector 1. Acquisition, Display, Archiving Transmitted x-rays through patient Exposed film Film processor Developer Fixer Wash Dry Gray Scale encoded on Film Intensifying Screens film x-rays → light Digital x-ray detector 2. Display Digital Pixel Digital to Analog 1. Acquisition Matrix Conversion Transmitted x-rays through patient Digital processing Analog to Digital Conversion Charge X-ray converter collection x-rays → electrons device 3. Archiving Analog versus Digital Spatial Resolution MTF of pixel aperture (DEL) 1 100 µm 0.8 0.6 200 µm 1000 µm 0.4 Modulation 0.2 0 01234567891011 Frequency (lp/mm) Sampling Detector Pitch Element, “DEL” 2 Characteristic Curve: Response of screen/film vs. digital detectors 5 Useless 4 10,000 Film-screen (400 speed) Digital 3 1,000 Overexposed Useless 2 100 Correctly exposed 1 10 intensity Relative Film Optical Density Film Optical Underexposed 0 1 0.01 0.1 1 10 100 Exposure, mR 20000 2000 200 20 2 Sensitivity (S) Analog versus digital detectors • Analog -

Digital Imaging Ethics
Digital Imaging Ethics It is strongly advised that EMS Users follow the MSA Policy on Digital Image Manipulation. "Ethical digital imaging requires that the original uncompressed image file be stored on archival media (e.g., CD-R) without any image manipulation or processing operation. All parameters of the production and acquisition of this file, as well as any subsequent processing steps, must be documented and reported to ensure reproducibility. Generally, acceptable (non-reportable) imaging operations include gamma correction, histogram stretching, and brightness and contrast adjustments. All other operations (such as Unsharp-Masking, Gaussian Blur, etc.) must be directly identified by the author as part of the experimental methodology. However, for diffraction data or any other image data that is used for subsequent quantification, all imaging operations must be reported." This policy was formulated by the Digital Image Processing & Ethics Group of the MSA Education Committee and was adopted as MSA policy at the Summer Council meeting August 2-3, 2003. Guidelines for the proper acquisition and manipulation of scientific digital images: (from Douglas W. Cromey, M.S. - Manager, Cellular Imaging Core Southwest Environmental Health Sciences Center, University of Arizona, Tucson, Arizona) See here for the original article These guidelines were written for life science imaging but are relevant to materials science microscopy as well. 1. Scientific digital images are data that can be compromised by inappropriate manipulations. Images are data arranged spatially in an XY matrix (or grid) and each individual element (pixel) has a numerical value that represents a grayscale or RGB intensity value. These data are a numerical sampling of the specimen as presented by the data acquisition system (e.g., microscope) to the sensor (e.g., CCD camera). -

Digital Imaging and Printing Selection 92 Image Manipulation Requires Selecting Either a Part of the Image Or the Entire Image to Make Changes
90 CHAPTER DIGITAL IMAGING and 08 PRINTING Towards a New AgeDesign a New Graphic Towards raphic designers work with visual images, either for Gprint media or for digital media. With the advent of 91 computers, most of the graphic designer’s work is being done using computers. From graphical point of view there is a vast difference between images on paper such as drawings, sketches or photographs and images that you see on the screen of the computers. Images that are created, manipulated and displayed using computers are called digital images. Digital images are different from images drawn or painted on paper in many ways. TYPES OF DIGITAL IMAGES There are two major categories of digital images: raster images and vector images. When images are stored in a computer in the form of a grid of basic picture elements called pixels, then these images are called raster images. The pixels contain the information about colour and brightness. Image-editing programmes can replace or modify the pixels to edit the image Digital Imaging and Imaging Printing Digital in various ways. The pixels can be modified in groups, or individually. There are sophisticated algorithms to achieve this. On the other hand vector images are stored as mathematical descriptions of the image in terms of lines, Bezier curves, and text instead of pixels. This is the main difference between raster and vector images. However, this difference is responsible for the development of two major categories of graphic technologies namely: Raster Graphics and Vector Graphics. Therefore, the image-editing software used by the practicing graphic designers falls under one of these categories. -

Art and Art History 1
Art and Art History 1 ART AND ART HISTORY art.as.miami.edu Dept. Codes: ART, ARH Educational Objectives The Department of Art and Art History provides facilities and instruction to serve the needs of the general student. The program fosters participation and appreciation in the visual arts for students with specialized interests and abilities preparing for careers in the production and interpretation of art and art history. Degree Programs The Department of Art and Art History offers two undergraduate degrees: • The Bachelor of Arts, with tracks in: • Art (General Study) • Art History • Studio Art • The Bachelor of Fine Arts in Studio Art, which allows for primary and secondary concentrations in: • Painting • Sculpture • Printmaking • Photography/Digital Imaging • Graphic Design/Multimedia • Ceramics The B.A. requires a minimum of 36 credit hours in the department with a grade of C or higher. The B. A. major is also required to have a minor outside the department. Minor requirements are specified by each department and are listed in the Bulletin. The B.F.A. requires a minimum of 72 credit hours in the department, a grade of C or higher in each course, a group exhibition and at least a 3.0 average in departmental courses. The B.F.A. major is not required to have a minor outside the department. Writing within the Discipline To satisfy the College of Arts and Sciences writing requirement in the discipline, students whose first major is art or art history must take at least one of the following courses for a writing credit: ARH 343: Modern Art, and/or ARH 344: Contemporary Art. -

(PPS) • CMOS Photodiode Active Pixel Sensor (APS) • Photoga
Lecture Notes 4 CMOS Image Sensors CMOS Passive Pixel Sensor (PPS) • Basic operation ◦ Charge to output voltage transfer function ◦ Readout speed ◦ CMOS Photodiode Active Pixel Sensor (APS) • Basic operation ◦ Charge to output voltage transfer function ◦ Readout speed ◦ Photogate and Pinned Diode APS • Multiplexed APS • EE 392B: CMOS Image Sensors 4-1 Introduction CMOS image sensors are fabricated in \standard" CMOS technologies • Their main advantage over CCDs is the ability to integrate analog and • digital circuits with the sensor Less chips used in imaging system ◦ Lower power dissipation ◦ Faster readout speeds ◦ More programmability ◦ New functionalities (high dynamic range, biometric, etc) ◦ But they generally have lower perofrmance than CCDs: • Standard CMOS technologies are not optimized for imaging ◦ More circuits result in more noise and fixed pattern noise ◦ In this lecture notes we discuss various CMOS imager architectures • In the following lecture notes we discuss fabrication and layout issues • EE 392B: CMOS Image Sensors 4-2 CMOS Image Sensor Architecture Word Pixel: Row Decoder Photodetector & Readout treansistors Bit Column Amplifiers/Caps Output Column Mux Readout performed by transferring one row at a time to the column • storage capacitors, then reading out the row, one (or more) pixel at a time, using the column decoder and multiplexer In many CMOS image sensor architectures, row integration times are • staggerred by the row/column readout time (scrolling shutter) EE 392B: CMOS Image Sensors 4-3 CMOS Image Sensor -

Digital Art & Design (DART)
Digital Art & Design (DART) 1 DART-170 Digital Video Editing 3 Units DIGITAL ART & DESIGN (DART) 108 hours activity; 108 hours total Introduction to non-linear editing on the computer. Includes historical DART-101 Graphic Design Foundations 3 Units development, digital video and audio formats, techniques and theory 108 hours activity; 108 hours total of editing, aspect ratios, organization of the edit, desktop environment, Graphic Design Foundations is an introductory course with emphasis on importing digital elements, project organization, video and audio files, the foundations of the Graphic Arts. Course content includes concept non-linear editing skills, applying transitions, designing titles, applying development, design processes, production, presentation, technical skills filters, digital and time line effects, importing graphics, mixing audio and in both traditional and digital media, and solving visual communication video elements, synchronize sound with video, and exporting digital video problems. Projects include lettering/typography and layout/composition. projects. Transfers to both UC/CSU Transfers to both UC/CSU DART-120 Intro to Digital Art & Graphic Design 3 Units 36 hours lecture; 54 hours lab; 90 hours total Recommended Preparation: Completion of ARTS-101 with a minimum grade of C. This course provides an introduction to visual design concepts and contemporary professional practices in graphic art using industry- standard software including Adobe Photoshop, Illustrator and InDesign. Transfers to both UC/CSU DART-125 Animation 3 Units 36 hours lecture; 54 hours lab; 90 hours total An introductory course in the basic principles and technology of animation. Both traditional and alternative animation styles will be covered with an emphasis on creating effective sequences appropriate for the subject or narrative. -
Digital Imaging Tutorial - Contents
Digital Imaging Tutorial - Contents 1. Preface 1. Basic Terminology 2. Selection 3. Conversion 4. Quality Control 5. Metadata Questions? 6. Technical Infrastructure A. Digitization Chain Table of Contents B. Image Creation C. File Management Using This Tutorial D. Delivery 7. Presentation Printing This Tutorial 8. Digital Preservation 9. Management 10. Continuing Education © 2000-2003 Cornell University Library/ Research Department http://www.library.cornell.edu/preservation/tutorial/contents.html [4/28/2003 2:27:14 PM] Digital Imaging Tutorial Questions? Use this form to send your questions and comments about the tutorial. Name: Complete Email Address: Question/Comment: © 2000-2003 Cornell University Library/Research Department http://www.library.cornell.edu/preservation/tutorial/questions.html [4/28/2003 2:27:16 PM] Digital Imaging Tutorial - Table of Contents PREFACE 5. METADATA 7. PRESENTATION 1. BASIC TERMINOLOGY definition introduction digital images types and functions formats/compression resolution creation web browsers pixel dimensions additional reading network bit depth scaling monitors dynamic range 6. TECHNICAL INFRASTRUCTURE file size image quality A. DIGITIZATION CHAIN compression guidelines introduction file formats additional reading components additional reading 8. DIGITAL PRESERVATION system integration 2. SELECTION definition B. IMAGE CREATION introduction challenges legal restrictions introduction technical strategies how scanners work organizational strategies other criteria selection policies scanner types additional reading image processing additional reading 9. MANAGEMENT . C FILE MANAGEMENT introduction 3.CONVERSION introduction project life cycle keeping track in-house vs. outsource image databases in-house facility introduction storage project budgets scanning factors storage types communication rich digital master storage needs project monitoring benchmarking D. DELIVERY looking beyond text additional reading stroke introduction continuous-tone networks 10. -

Carl Djerassi 1923‐2015
SenD#7009 MEMORIAL RESOLUTION CARL DJERASSI 1923‐2015 Carl Djerassi, Professor Emeritus of Chemistry, novelist, playwright, patron of the arts and pioneer in the production of the oral contraceptive pill, died in his home in San Francisco on January 30. He was 91 years old. Djerassi was born in Vienna in 1923 to Samuel Djerassi and Alice Friedmann. He lived for periods in Sofia, Bulgaria and Vienna but with the rising Nazi threat, moved with his mother to the United States, arriving nearly penniless in 1939. Djerassi graduated at the age of 18 from Kenyon College where in his own words, he “became a chemist”. He earned a Ph.D. in chemistry from Wisconsin in 1945 and subsequently worked at CIBA, developing one of the first commercial antihistamines (Pyribenzamine). In 1949, Djerassi became associate director of research at Syntex in Mexico City. His research on steroids led to the synthesis of norethisterone and the first birth‐control pill, studies that have since transformed science and society. Djerassi joined the faculty at Wayne State University in 1952. In 1959, he joined the faculty at Stanford while for a time serving also as president of Syntex Research. In 1968, he founded Zoecon, a company directed at hormonal control of insect pests. Djerassi published over 1200 scientific manuscripts, encompassing seminal contributions to tools for structure elucidation including mass spectrometry, magnetic circular dichroism and optical rotatory dispersion. He also made pioneering contributions to our understanding of the biosynthesis of marine natural products. With Nobel Laureate Joshua Lederberg and computer scientist Edward Feigenbaum, he also devised a computer program called DENDRAL, one of the earliest uses of artificial intelligence for structure elucidation.