Loss of the SWI/SNF-Atpase Subunit Members SMARCF1 (ARID1A)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

SMARCB1/INI1 Genetic Inactivation Is Responsible for Tumorigenic Properties of Epithelioid Sarcoma Cell Line VAESBJ
Published OnlineFirst April 10, 2013; DOI: 10.1158/1535-7163.MCT-13-0005 Molecular Cancer Cancer Therapeutics Insights Therapeutics SMARCB1/INI1 Genetic Inactivation Is Responsible for Tumorigenic Properties of Epithelioid Sarcoma Cell Line VAESBJ Monica Brenca1, Sabrina Rossi3, Erica Lorenzetto1, Elena Piccinin1, Sara Piccinin1, Francesca Maria Rossi2, Alberto Giuliano1, Angelo Paolo Dei Tos3, Roberta Maestro1, and Piergiorgio Modena1 Abstract Epithelioid sarcoma is a rare soft tissue neoplasm that usually arises in the distal extremities of young adults. Epithelioid sarcoma presents a high rate of recurrences and metastases and frequently poses diagnostic dilemmas. We previously reported loss of tumor suppressor SMARCB1 protein expression and SMARCB1 gene deletion in the majority of epithelioid sarcoma cases. Unfortunately, no appropriate preclinical models of such genetic alteration in epithelioid sarcoma are available. In the present report, we identified lack of SMARCB1 protein due to a homozygous deletion of exon 1 and upstream regulatory region in epithelioid sarcoma cell line VAESBJ. Restoration of SMARCB1 expression significantly affected VAESBJ cell proliferation, anchorage-independent growth, and cell migration properties, thus supporting the causative role of SMARCB1 loss in epithelioid sarcoma pathogenesis. We investigated the translational relevance of this genetic back- ground in epithelioid sarcoma and showed that SMARCB1 ectopic expression significantly augmented VAESBJ sensitivity to gamma irradiation and acted synergistically with flavopiridol treatment. In VAESBJ, both activated ERBB1/EGFR and HGFR/MET impinged on AKT and ERK phosphorylation. We showed a synergistic effect of combined inhibition of these 2 receptor tyrosine kinases using selective small-molecule inhibitors on cell proliferation. These observations provide definitive support to the role of SMARCB1 inactivation in the pathogenesis of epithelioid sarcoma and disclose novel clues to therapeutic approaches tailored to SMARCB1-negative epithelioid sarcoma. -

Integrase Interactor 1 (INI-1) Deficient Renal Cell Carcinoma
Open Access Case Report DOI: 10.7759/cureus.13082 Integrase Interactor 1 (INI-1) Deficient Renal Cell Carcinoma Manpreet Singh 1 , Harkirat Singh 1 , Benjamin Hambro 1 , Jasleen Kaur 1 , Ravi Rao 2 1. Internal Medicine, St Agnes Medical Center, Fresno, USA 2. Hematology and Oncology, St Agnes Medical Center, Fresno, USA Corresponding author: Manpreet Singh, [email protected] Abstract Members of the SWItch/sucrose nonfermentable (SWI-SNF) family, including SWI/SNF related, matrix- associated, actin-dependent regulator of chromatin, subfamily A, member 4 (SMARCA4), SWI/SNF related, matrix‐associated, actin‐dependent regulator of chromatin, subfamily B member 1 (SMARCB1)/integrase interactor 1 (INI-1) are known tumor suppressor genes. Interactions between SMARCB1/INI-1 and key protein components in various cellular pathways are related to tumor progression and proliferation. SMARCB1/INI-1 protein was undetectable in rhabdoid tumor cells, whereas non-tumorous cells express the SMARCB1/INI-1 genes. Germline and sporadic mutations of several genes encoding for proteins in this complex are known to cause a spectrum of cancers, usually with sarcomatoid features which include a very aggressive renal medullary carcinoma. We report a case of a 29-year-old male who presented with SMARCA4 deficient renal tumor with a very aggressive clinical behavior which ultimately led to his death. Categories: Nephrology, Oncology Keywords: renal medullary carcinoma, ini-1, renal cell carcinoma, swi/snf, smarcb1, integrase interactor 1 Introduction Renal medullary carcinoma is a rare and very aggressive malignancy affecting young adults with rare cases in patients with sickle cell disease or trait [1]. The tumor arises predominantly in the renal medulla and exhibits a variety of growth patterns including reticular, solid, tubular, trabecular, cribriform, sarcomatoid, and micropapillary [1]. -

Loss of Integrase Interactor 1 (INI1) Expression in a Subset of Differentiated Thyroid Cancer
diagnostics Article Loss of Integrase Interactor 1 (INI1) Expression in a Subset of Differentiated Thyroid Cancer Kung-Chen Ho 1, Jie-Jen Lee 1, Chi-Hsin Lin 2,3, Ching-Hsiang Leung 4 and Shih-Ping Cheng 1,5,* 1 Department of Surgery, MacKay Memorial Hospital and Mackay Medical College, Taipei 104215, Taiwan; [email protected] (K.-C.H.); [email protected] (J.-J.L.) 2 Department of Medical Research, MacKay Memorial Hospital, Taipei 104215, Taiwan; [email protected] 3 Department of Bioscience Technology, Chung Yuan Christian University, Taoyuan City 320314, Taiwan 4 Division of Endocrinology and Metabolism, Department of Internal Medicine, MacKay Memorial Hospital and Mackay Medical College, Taipei 104215, Taiwan; [email protected] 5 Department of Pharmacology, School of Medicine, College of Medicine, Taipei Medical University, Taipei 110301, Taiwan * Correspondence: [email protected]; Tel.: +886-2-2543-3535 Received: 19 April 2020; Accepted: 2 May 2020; Published: 5 May 2020 Abstract: Alterations in the switching defective/sucrose non-fermenting (SWI/SNF) chromatin-remodeling complex are enriched in advanced thyroid cancer. Integrase interactor 1 (INI1), encoded by the SMARCB1 gene on the long arm of chromosome 22, is one of the core subunits of the SWI/SNF complex. INI1 immunohistochemistry is frequently used for the diagnosis of malignant rhabdoid neoplasms. In the present study, we found normal and benign thyroid tissues generally had diffusely intense nuclear immunostaining. Loss of INI1 immunohistochemical expression was observed in 8% of papillary thyroid cancer and 30% of follicular thyroid cancer. Furthermore, loss of INI1 expression was associated with extrathyroidal extension (p < 0.001) and lymph node metastasis (p = 0.038). -

Renal Medullary Carcinomas Depend Upon SMARCB1 Loss And
RESEARCH ARTICLE Renal medullary carcinomas depend upon SMARCB1 loss and are sensitive to proteasome inhibition Andrew L Hong1,2,3, Yuen-Yi Tseng3, Jeremiah A Wala3, Won-Jun Kim2, Bryan D Kynnap2, Mihir B Doshi3, Guillaume Kugener3, Gabriel J Sandoval2,3, Thomas P Howard2, Ji Li2, Xiaoping Yang3, Michelle Tillgren2, Mahmhoud Ghandi3, Abeer Sayeed3, Rebecca Deasy3, Abigail Ward1,2, Brian McSteen4, Katherine M Labella2, Paula Keskula3, Adam Tracy3, Cora Connor5, Catherine M Clinton1,2, Alanna J Church1, Brian D Crompton1,2,3, Katherine A Janeway1,2, Barbara Van Hare4, David Sandak4, Ole Gjoerup2, Pratiti Bandopadhayay1,2,3, Paul A Clemons3, Stuart L Schreiber3, David E Root3, Prafulla C Gokhale2, Susan N Chi1,2, Elizabeth A Mullen1,2, Charles WM Roberts6, Cigall Kadoch2,3, Rameen Beroukhim2,3,7, Keith L Ligon2,3,7, Jesse S Boehm3, William C Hahn2,3,7* 1Boston Children’s Hospital, Boston, United States; 2Dana-Farber Cancer Institute, Boston, United States; 3Broad Institute of Harvard and MIT, Cambridge, United States; 4Rare Cancer Research Foundation, Durham, United States; 5RMC Support, North Charleston, United States; 6St. Jude Children’s Research Hospital, Memphis, United States; 7Brigham and Women’s Hospital, Boston, United States Abstract Renal medullary carcinoma (RMC) is a rare and deadly kidney cancer in patients of African descent with sickle cell trait. We have developed faithful patient-derived RMC models and using whole-genome sequencing, we identified loss-of-function intronic fusion events in one SMARCB1 allele with concurrent loss of the other allele. Biochemical and functional characterization of these models revealed that RMC requires the loss of SMARCB1 for survival. -

Regulation of P27kip1 and P57kip2 Functions by Natural Polyphenols
biomolecules Review Regulation of p27Kip1 and p57Kip2 Functions by Natural Polyphenols Gian Luigi Russo 1,* , Emanuela Stampone 2 , Carmen Cervellera 1 and Adriana Borriello 2,* 1 National Research Council, Institute of Food Sciences, 83100 Avellino, Italy; [email protected] 2 Department of Precision Medicine, University of Campania “Luigi Vanvitelli”, 81031 Napoli, Italy; [email protected] * Correspondence: [email protected] (G.L.R.); [email protected] (A.B.); Tel.: +39-0825-299-331 (G.L.R.) Received: 31 July 2020; Accepted: 9 September 2020; Published: 13 September 2020 Abstract: In numerous instances, the fate of a single cell not only represents its peculiar outcome but also contributes to the overall status of an organism. In turn, the cell division cycle and its control strongly influence cell destiny, playing a critical role in targeting it towards a specific phenotype. Several factors participate in the control of growth, and among them, p27Kip1 and p57Kip2, two proteins modulating various transitions of the cell cycle, appear to play key functions. In this review, the major features of p27 and p57 will be described, focusing, in particular, on their recently identified roles not directly correlated with cell cycle modulation. Then, their possible roles as molecular effectors of polyphenols’ activities will be discussed. Polyphenols represent a large family of natural bioactive molecules that have been demonstrated to exhibit promising protective activities against several human diseases. Their use has also been proposed in association with classical therapies for improving their clinical effects and for diminishing their negative side activities. The importance of p27Kip1 and p57Kip2 in polyphenols’ cellular effects will be discussed with the aim of identifying novel therapeutic strategies for the treatment of important human diseases, such as cancers, characterized by an altered control of growth. -

Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development
International Journal of Molecular Sciences Article Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development Nashwa El Hadidy 1 and Vladimir N. Uversky 1,2,* 1 Department of Molecular Medicine, Morsani College of Medicine, University of South Florida, 12901 Bruce B. Downs Blvd. MDC07, Tampa, FL 33612, USA; [email protected] 2 Laboratory of New Methods in Biology, Institute for Biological Instrumentation of the Russian Academy of Sciences, Federal Research Center “Pushchino Scientific Center for Biological Research of the Russian Academy of Sciences”, Pushchino, 142290 Moscow Region, Russia * Correspondence: [email protected]; Tel.: +1-813-974-5816; Fax: +1-813-974-7357 Received: 20 September 2019; Accepted: 21 October 2019; Published: 23 October 2019 Abstract: The two-meter-long DNA is compressed into chromatin in the nucleus of every cell, which serves as a significant barrier to transcription. Therefore, for processes such as replication and transcription to occur, the highly compacted chromatin must be relaxed, and the processes required for chromatin reorganization for the aim of replication or transcription are controlled by ATP-dependent nucleosome remodelers. One of the most highly studied remodelers of this kind is the BRG1- or BRM-associated factor complex (BAF complex, also known as SWItch/sucrose non-fermentable (SWI/SNF) complex), which is crucial for the regulation of gene expression and differentiation in eukaryotes. Chromatin remodeling complex BAF is characterized by a highly polymorphic structure, containing from four to 17 subunits encoded by 29 genes. The aim of this paper is to provide an overview of the role of BAF complex in chromatin remodeling and also to use literature mining and a set of computational and bioinformatics tools to analyze structural properties, intrinsic disorder predisposition, and functionalities of its subunits, along with the description of the relations of different BAF complex subunits to the pathogenesis of various human diseases. -

Global Chromatin Changes Resulting from Single-Gene Inactivation—The Role of SMARCB1 in Malignant Rhabdoid Tumor
cancers Article Global Chromatin Changes Resulting from Single-Gene Inactivation—The Role of SMARCB1 in Malignant Rhabdoid Tumor Colin Kenny 1, Elaine O’Meara 2, Mevlüt Ula¸s 2 , Karsten Hokamp 3 and Maureen J. O’Sullivan 1,2,4,* 1 School of Medicine, Trinity College, University of Dublin, Dublin 2, Ireland; [email protected] 2 The National Children’s Research Centre, O’Sullivan Research Laboratory, Oncology Division, Gate 5, Children’s Health Ireland at Crumlin, D12N512 Dublin, Ireland; [email protected] (E.O.); [email protected] (M.U.) 3 School of Genetics and Microbiology, Trinity College, University of Dublin, Dublin 2, Ireland; [email protected] 4 Histology Laboratory, Pathology Department, Children’s Health Ireland at Crumlin, D12N512 Dublin, Ireland * Correspondence: [email protected] Simple Summary: Malignant rhabdoid tumors (MRT), one of the most lethal, treatment-resistant human cancers, arises in young children within brain, kidney, liver and/or soft tissues. Generally, cancer arises in older adults, and results from multiple significant changes (mutations) accumulating in the genetic blueprint (DNA) of a person’s tissues. This blueprint is composed of a 4-letter alphabet. Together, the multiple significant changes in the blueprint then allow a cell to go “out of control”, becoming a cancer cell. The striking thing about MRT is that it has only a single spelling change, so that mutation must be very powerful to lead to such a lethal cancer. Using a model system that we developed, we show herein how this single mutation alters how the whole of the DNA is arranged, Citation: Kenny, C.; O’Meara, E.; Ula¸s,M.; Hokamp, K.; O’Sullivan, thereby having its profound and lethal effects. -
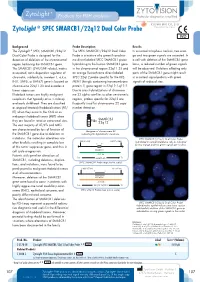
Zytolight ® SPEC SMARCB1/22Q12 Dual Color Probe
ZytoLight ® SPEC SMARCB1/22q12 Dual Color Probe Background Probe Description Results The ZytoLight ® SPEC SMARCB1/22q12 The SPEC SMARCB1/22q12 Dual Color In a normal interphase nucleus, two oran- Dual Color Probe is designed for the Probe is a mixture of a green fluorochro- ge and two green signals are expected. In detection of deletions of the chromosomal me direct labeled SPEC SMARCB1 probe a cell with deletion of the SMARCB1 gene region harboring the SMARCB1 gene. hybridizing to the human SMARCB1 gene locus, a reduced number of green signals The SMARCB1 (SWI/SNF related, matrix in the chromosomal region 22q11.23 and will be observed. Deletions affecting only associated, actin dependent regulator of an orange fluorochrome direct labeled parts of the SMARCB1 gene might result chromatin, subfamily b, member 1, a.k.a. SPEC 22q12 probe specific for the KRE- in a normal signal pattern with green INI1, SNF5, or BAF47) gene is located on MEN1 (kringle containing transmembrane signals of reduced size. chromosome 22q11.23 and encodes a protein 1) gene region in 22q12.1-q12.2. tumor suppressor. Due to cross-hybridizations of chromoso- Rhabdoid tumors are highly malignant me 22 alpha satellites to other centromeric neoplasms that typically arise in infancy regions, probes specific for 22q12 are and early childhood. They are classified frequently used for chromosome 22 copy as atypical teratoid/rhabdoid tumors (AT/ number detection. RT) when they occur in the CNS or as malignant rhabdoid tumors (MRT) when they are found in renal or extra-renal sites. SMARCB1 22q12 The vast majority of AT/RTs and MRTs are characterized by loss of function of Ideogram of chromosome 22 the SMARCB1 gene due to deletions or indicating the hybridization locations. -

ARID1A Mutations in Cancer: Another Epigenetic Tumor Suppressor?
Published OnlineFirst December 3, 2012; DOI: 10.1158/2159-8290.CD-12-0361 MINI REVIEW ARID1A Mutations in Cancer: Another Epigenetic Tumor Suppressor? Jennifer N. Wu and Charles W.M. Roberts ABSTRACT Although disordered chromatin organization has long been recognized as a fea- ture of cancer, the molecular underpinnings of chromatin structure, epigenetic regulation, and their relationships to transcription are only beginning to be understood. Cancer genome sequencing studies have revealed a novel theme: frequent mutation of epigenetic regulators. Among these, the ARID1A/BAF250A subunit of the SWI/SNF (BRG1-associated factors) chromatin remod- eling complex has emerged as recurrently mutated in a broad array of tumor types. We review the genomic and functional data supporting classifi cation of ARID1A as a tumor suppressor. Signifi cance: Mutations in chromatin remodeling complex genes are increasingly recognized in many cancer types. However, the mechanisms by which chromatin remodeling complexes contribute to gene expression and the cancer phenotype are poorly understood. Understanding how mutation of chroma- tin remodelers facilitates transformation may offer the potential for development and implementation of novel therapies for cancer. Cancer Discov; 3(1); 35–43. ©2012 AACR. INTRODUCTION risk of developing malignant rhabdoid tumors at an especially young age. Epigenetic regulators impose upon the genetic code a Subsequent to these discoveries, implementation of next- chromatin structure characterized by chromatin accessibil- generation sequencing technologies have revealed frequent ity, nucleosome position, and histone modifi cations. These and recurrent mutations in a wide variety of epigenetic features of chromatin structure modulate gene expres- modulators, including mediators of DNA methylation (e.g., sion, thereby affecting both the identity and function of DNMT3a) and covalent modifi ers of histones (e.g., MLL, MLL3, cells. -

Low-Dose Histone Deacetylase Inhibitor Treatment Leads to Tumor
Published OnlineFirst February 26, 2016; DOI: 10.1158/1078-0432.CCR-15-2260 Cancer Therapy: Preclinical Clinical Cancer Research Low-Dose Histone Deacetylase Inhibitor Treatment Leads to Tumor Growth Arrest and Multi-Lineage Differentiation of Malignant Rhabdoid Tumors Andrea Muscat1,2, Dean Popovski3,4,W. Samantha N. Jayasekara3,4, Fernando J. Rossello5,6, Melissa Ferguson1,2, Kieren D. Marini3,4, Muhammad Alamgeer3,7, Elizabeth M. Algar3,4, Peter Downie8,9, D. Neil Watkins3,4,10, Jason E. Cain3,4, and David M. Ashley1,2 Abstract Purpose: Malignant rhabdoid tumor (MRT) and atypical Results: Sustained low-dose panobinostat (LBH589) treat- teratoid rhabdoid tumors (ATRT) are rare aggressive undiffer- ment led to changes in cellular morphology associated with a entiated tumors primarily affecting the kidney and CNS of marked increase in the induction of neural, renal, and osteoblast infants and young children. MRTarealmostexclusivelychar- differentiation pathways. Genome-wide transcriptional profiling acterized by homozygous deletion or inactivation of the chro- highlighted differential gene expression supporting multilineage matin remodeling gene SMARCB1. SMARCB1 protein loss leads differentiation. Using mouse xenograft models, sustained low- to direct impairment of chromatin remodeling and we have dose LBH589 treatment caused tumor growth arrest associated previously reported a role for this protein in histone acetyla- with tumor calcification detectable by X-ray imaging. Histological tion. This provided the rationale for investigating the thera- analysis of LBH589-treated tumors revealed significant regions of peutic potential of histone deactylase inhibitors (HDACi) in ossification, confirmed by Alizarin Red staining. Immunohisto- MRT. chemical analysis showed increased TUJ1 and PAX2 staining Experimental Design: Whereas previously HDACis have been suggestive of neuronal and renal differentiation, respectively. -

The Chromatin Remodelling Component SMARCB1/INI1
Pancione et al. Journal of Translational Medicine 2013, 11:297 http://www.translational-medicine.com/content/11/1/297 RESEARCH Open Access The chromatin remodelling component SMARCB1/INI1 influences the metastatic behavior of colorectal cancer through a gene signature mapping to chromosome 22 Massimo Pancione1*†, Andrea Remo2†, Caterina Zanella2, Lina Sabatino1, Arturo Di Blasi3, Carmelo Laudanna1, Laura Astati2, Michele Rocco4, Delfina Bifano4, Paolo Piacentini2, Laura Pavan2, Alberto Purgato2, Filippo Greco2, Alberto Talamini2, Andrea Bonetti2, Michele Ceccarelli1, Roberto Vendraminelli2, Erminia Manfrin5 and Vittorio Colantuoni1* Background: INI1 (Integrase interactor 1), also known as SMARCB1, is the most studied subunit of chromatin remodelling complexes. Its role in colorectal tumorigenesis is not known. Methods: We examined SMARCB1/INI1 protein expression in 134 cases of colorectal cancer (CRC) and 60 matched normal mucosa by using tissue microarrays and western blot and categorized the results according to mismatch repair status (MMR), CpG island methylator phenotype, biomarkers of tumor differentiation CDX2, CK20, vimentin and p53. We validated results in two independent data sets and in cultured CRC cell lines. Results: Herein, we show that negative SMARCB1/INI1 expression (11% of CRCs) associates with loss of CDX2, poor differentiation, liver metastasis and shorter patients’ survival regardless of the MMR status or tumor stage. Unexpectedly, even CRCs displaying diffuse nuclear INI1 staining (33%) show an adverse prognosis and vimentin over-expression, in comparison with the low expressing group (56%). The negative association of SMARCB1/INI1-lack of expression with a metastatic behavior is enhanced by the TP53 status. By interrogating global gene expression from two independent cohorts of 226 and 146 patients, weconfirmtheprognosticresults and identify a gene signature characterized by SMARCB1/INI1 deregulation. -

Associations Between Human Disease Genes and Overlapping Gene Groups and Multiple Amino Acid Runs
Associations between human disease genes and overlapping gene groups and multiple amino acid runs Samuel Karlin*†, Chingfer Chen*, Andrew J. Gentles*, and Michael Cleary‡ Departments of *Mathematics and ‡Pathology, Stanford University, Stanford, CA 94305 Contributed by Samuel Karlin, October 30, 2002 Overlapping gene groups (OGGs) arise when exons of one gene are Chr 22. Such rearrangements may be mediated by recombination contained within the introns of another. Typically, the two over- events based on region-specific low copy repeats. The DGS lapping genes are encoded on opposite DNA strands. OGGs are region of 22q11.2 is particularly rich with segmental duplications, often associated with specific disease phenotypes. In this report, which can induce deletions, translocations, and genomic insta- we identify genes with OGG architecture and genes encoding bility (4). There are several anomalous sequence features asso- multiple long amino acid runs and examine their relations to ciated with OGGs, including Alu sequences intersecting exons, diseases. OGGs appear to be susceptible to genomic rearrange- pseudogenes occupying introns, and single-exon (intronless) ments as happens commonly with the loci of the DiGeorge syn- genes that often result from a processed multiexon gene. drome on human chromosome 22. We also examine the degree of At least 28 genes in Chr 21 are related to diseases, as conservation of OGGs between human and mouse. Our analyses characterized in the GeneCards database (5), as are 64 genes in suggest that (i) a high proportion of genes in OGG regions are Chr 22. Specific disorders that have been mapped to genes on disease-associated, (ii) genomic rearrangements are likely to occur Chr 21 and that involve OGG structures include: amyotrophic within OGGs, possibly as a consequence of anomalous sequence lateral sclerosis (ALS, Lou Gehrig’s disease), linked to the features prevalent in these regions, and (iii) multiple amino acid GRIK1 ionotrophic kainate 1 glutamate receptor gene at 21q22 runs are also frequently associated with pathologies.