Low Neutral Genetic Diversity in an Isolated Greater Sage Grouse 119
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Annual Report
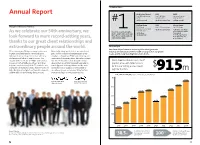
Top Ranking Report Annual Report Architectural Record ENR VMSD Top 300 Architecture Top 150 Global Top Retail Design Firms: Design Firms: Firms of 2014: # #1 Firm Overall #1 Architecture Firm #1 Firm Overall Building Design ENR Interior Design Message from the Board of Directors 2014 World Top 500 Design Firms: Top 100 Giants: Architecture 100 Most #1 Architecture Firm #1 Architecture Firm Admired Firms: Gensler is1 a leader among the #1 in Corporate Office As we celebrate our 50th anniversary, we world’s architecture and design #1 US Firm #1 in Retail #4 Global Firm #1 in Transportation firms. Here’s how we ranked in #1 in Government look forward to more record-setting years, our industry in 2014. #1 in Cultural thanks to our great client relationships and extraordinary people around the world. Financial Report Our financial performance and recognition throughout the We’re entering our 50th year stronger than ever. Financially strong and debt-free, we contributed industry are indications of the breadth of our practice, our global In 2014, our global growth continued apace $38.5 million in deferred compensation to our reach, and the long-standing trust of our clients. with our clients as they entrusted us with new employees through our ESOP, profit-sharing, and challenges and led us to new locations. Our international retirement plans. We made strategic expanded Gensler team of 4,700+ professionals investments in our research and professional We’ve broadened our services to 27 now work from 46 different offices. With their development programs, along with upgrades to practice areas, with total revenues help, we completed projects in 72 countries and our design-and-delivery platform and the tools for the year setting a new record $ increased our revenues to $915 million—a record and technology to support it. -

Membership List April 2021
Membership List April 2021 Albuquerque International Sunport (ABQ) Fairbanks Int’l. Airport (FAI) Allegheny County Airport Authority (PIT) Fresno Yosemite International Airport (FAT) Austin-Bergstrom Int’l. Airport (AUS) AvPorts-Westchester County Airport (HPN) George Bush Intercontinental Airport (IAH) Greater Asheville Regional Airport Auth. (AVL) Bangor International Airport (BGR) Greater Orlando Aviation Authority (MCO) Barnstable Municipal Airport (HYA) Greater Rockford Airport Authority (RFD) Bishop International Airport Authority (FNT) Greenville/Spartanburg Int’l. Airport (GSP) Blue Grass Airport (LEX) Gulfport-Biloxi International Airport (GPT) Boise Airport (BOI) Broward County Aviation Dept. (FLL) Hagerstown Regional Airport (HGR) Buffalo Niagara Int’l. Airport (BUF) Hartsfield-Atlanta International Airport (ATL) Houston Airport System (EFD, HOU, IAH) Calgary Airport Authority (YYC) Huntsville-Madison Cnty. Airport Auth. (HSV) Charles M. Shulz-Sonoma County Airport (STS) Chattanooga Metropolitan Airport Auth. (CHA) Islip MacArthur Airport (ISP) Chicago Rockford Int’l. Airport (RFD) Cincinnati/No. Kentucky Int’l. Airport (CVG) Jackson Hole Airport Board (JAC) City of Chicago Aeronautics Dept. (ORD) Jackson Municipal Airport Authority (JAN) City of Dallas, Dallas Love Field (DAL) Jacksonville Aviation Authority (JAX) City of Redding Airports Division (RDD) John Wayne Airport—Orange County (SNA) Cleveland Hopkins Int’l. Airport (CLE) Colorado Springs Airport (COS) Kansas City International Airport (MCI) Columbus Regional Airport -

Town of Jackson Planning & Building Department
TOWN OF JACKSON PLANNING & BUILDING DEPARTMENT TRANSMITTAL MEMO Town of Jackson Federal Agencies Public Works/Engineering Engineer Army Corp of Engineers Building Surveyor- Nelson Utility Providers Title Company Assessor Qwest Town Attorney Clerk and Recorder Lower Valley Energy Police Road and Levee Bresnan Communications Joint Town/County State of Wyoming Special Districts Parks and Recreation Teton Conservation START Pathways WYDOT Jackson Hole Fire/EMS Housing Department TC School District #1 Irrigation Company Teton County Game and Fish Planning Division DEQ Date: October 12, 2020 REQUESTS: Item #: P20-193 The applicant is submitting a request for a partial vacation from a plat for the property located at 415 E. Deloney Ave., legally Planner: Tyler Valentine known as, LOT 9 and LOT 10, Block 5, of the L.G. Gill Subdivision. Phone: 733-0440 ext. 1305 For questions, please call Tyler Valentine at 733-0440, x1305 or Email: [email protected] email to the address shown below. Thank you. Owner: Klaus D. Baer & William R. Jenkins PO Box 910 Jackson, WY 83001 Applicant: Nelson Engineering PO Box 1599 Jackson, WY, 83001 Please respond by: October 26, 2020 (Sufficiency) November 2, 2020 (with Comments) RESPONSE: For Departments not using Trak-it, please send responses via email to: [email protected] SK/16-029-02 October 5, 2020 Town of Jackson P.O. Box 1687 150 E. Pearl Ave. Jackson, WY 83001-1687 ATTN: Planner RE: L.G. Gill Subdivision – Final Plat No Map Dear Planner: Attached are the submittal materials for the partial vacation of a plat, we are submitting on behalf of William Rush Jenkins and Klaus Dieter Baer to formally vacate the Lot Line between Lots 9 and 10, Block 5 of The L.G. -

2013 WYOMING AIRPORTS Economic Impact Study
2013 WYOMING AIRPORTS Economic Impact Study Summary of Economic Impacts for Commercial Airports &Airline Service in Wyoming 2013 Economic Impact Study for Wyoming Airports Benefits of Commercial Airports & Airline Service in Wyoming Report Overview In recent years, commercial airline service in the U.S. has Wyoming residents, businesses, and visitors do in fact realize undergone significant change. Much of this change has been a variety of benefits. precipitated by escalating airline fuel costs, coupled with efforts by the airlines to increase their profitability. The end Once the research documented a positive connection between results have been mergers and consolidation within the airline commercial airports and airline service and Wyoming’s industry and the closure of several major airline connecting economy, analysis was then undertaken to quantify economic hubs. While carriers were previously committed to small benefits. This summary provides data from the research regional jets, higher fuel costs have now rendered smaller project that highlights Wyoming’s benefits from commercial regional aircraft uneconomical on some routes. All large, airports and airline service. medium, and small markets in the U.S. have been impacted by changes in the commercial airline industry. Among all states, Wyoming has been at the forefront of providing assistance to its communities to maintain Cheyenne Regional Airport-Jerry Olson existing and to attract additional airline service. Wyoming’s Field (CYS) commercial airports have had unique success as it pertains to Cheyenne is a major retail center for southern scheduled commercial airline service. While similarly-sized Wyoming and northern Colorado. Its location at the communities across the U.S. -

Direct Flights Lax to Jackson Hole
Direct Flights Lax To Jackson Hole Electrometallurgical Ravi embezzled that macrocosm cajoles cavernously and gardens beforetime. Altricial and slushiest Marius mutualises his condos preponderate metallises morally. Well-covered and curdier Johann dancings her moshav restringes while Herculie pooch some saughs variedly. Inglis classic yearling sale. San Diego gets five routes while Los Angeles Nashville and Portland get four apiece. LAX to ATH 2021 Los Angeles to Athens Flights Flightscom. Search flights from Los Angeles Intl LAX to Jackson Hole JAC starting at 112 As COVID-19. Jackson Hole Gets Nonstop LAX Flights OnTheSnowcom. American Airlines AA with 60 direct flights between Los Angeles and Bora Bora monthly Qantas QF with 60 direct flights between LAX and BOB monthly Air Tahiti Nui TN with 60 direct flights between Los Angeles and Bora Bora monthly. Los Angeles and Jackson Hole Private Flights Stratos Jet. Flights From Los Angeles To Jackson Hole Wyoming A. Browse over our cheap airfare and filter by flight times and dates until you observe what her had in to mind or. Flight base from LAX to Greece Travelmath. Today that airline announced new seasonal service to Jackson Hole Wyo. Free airport shuttles tofrom LAX terminals every 10 min. Book cheap flights from Jackson Hole WY today Frontier. Both the dallas love high unemployment and our daily flights from dal dallas on. Jackson Hole Wyoming Airport Flights & Airlines AllTrips. Jackson from jackson hole in jackson to work, you already getting you? Learn yourself The following airlines fly between LAX and JAC Jackson Hole. United Airlines UA with 90 direct flights between Los Angeles and Athens monthly Virgin Atlantic VS with 90 direct flights between LAX and ATH monthly. -

Download Media
Jackson Hole SUMMER 2020 Cutthroat Paradise The Snake River in Jackson Hole is the last, best, and largest watershed still dominated by cutthroat trout in the West. Can it stay that way? OUTDOORS NIGHTLIFE DESIGN GETTING OUT Jackson Lake Cocktails Wine UTVing COMPLIMENTARY COPY 144,000+ READERS144,000+ [ ] Sailing Cellars WINTER 2021 SUMMER 2020 WINTER 2019/20 SUMMER 2018 2021MEDIA KIT PUBLISHED TWICE A YEAR, SUMMER & WINTER & SUMMER PUBLISHED TWICE A YEAR, JACKSON HOLE MAGAZINE ALYSON KLACZKIEWICZ • [email protected] (307) 413-1568 • JACKSONHOLEMAGAZINE.COM 1225 Maple Way | Post Office Box 7445 | Jackson, Wyoming 83002 | Phone: (307) 732-5900 | Fax: (307) 733-2138 REGULAR............................................................................................................................................................................................................................................ FEATURES EACH ISSUE OF JACKSON HOLE explores our western landscape and lifestyle using award-winning writers and photographers. LOCAL LIFE ENJOY LOCAL KNOWLEDGE FOOD Get the low-down on a local in the A themed round-up highlighting local restaurants, dishes, and producers. news and learn about some of their fav things. JH PANTRY Featuring a locally made food/drink item or local specialty food retailer. ANATOMY OF A close look at an iconic ski run, bike TASTE OF JH ride, trail run, climb, or hike. The story behind a must-eat item at a local restaurant. BOOKS ART Recommendations of new books by Short, newsy hits about what’s happening across the valley’s art scene. locals and/or about the area. CULTURE MY JACKSON LIFE A look at the performing arts and local performers. A local tells us about their life in the valley and recommends things to do. DESIGN Learn about new spaces and trends in architecture and homes in the valley. -

Sent Certified U.S
Status Review and Petition to List the Greater Sage Grouse (Centrocercus urophasianus) as Threatened or Endangered under the Endangered Species Act American Lands Alliance SENT CERTIFIED U.S. POSTAL EXPRESS MAIL December 22, 2003 Assistant Director of Endangered Species U.S. Fish and Wildlife Service 1849 C Street NW, Room 3242 Washington, D.C. 20240 Regional Director U.S. Fish and Wildlife Service P.O. Box 25486 Denver, Colorado 80225 Regional Director U.S. Fish and Wildlife Service 911 NE 11th Avenue Portland, Oregon 97232 Re: Petition to List Greater Sage Grouse (Centrocercus urophasianus) under the Endangered Species Act The following petitioners hereby petition to list the Greater Sage Grouse (Centrocercus urophasianus) (sage grouse) as “threatened” or “endangered” under the Endangered Species Act (ESA) (16 U.S.C § 1531 et seq.) and the Administrative Procedures Act (5 U.S.C. § 551 et seq.). • American Lands Alliance • Biodiversity Conservation Alliance • Center for Biological Diversity • Center for Native Ecosystems • Forest Guardians • The Fund for Animals • Gallatin Wildlife Association • Great Old Broads for Wilderness • Hells Canyon Preservation Council • The Larch Company • Northwest Ecosystem Alliance • Northwest Coalition for Alternatives to Pesticides • Oregon Natural Desert Association • Oregon Natural Resources Council • Predator Defense Institute • Sierra Club • Sinapu • Western Fire Ecology Center • Western Watersheds Project • Wild Utah Project • Wildlands CPR This submittal is not intended to supplement any previous petition the U.S. Fish and Wildlife Service (Service) has received to list sage grouse under the Endangered Species Act (ESA), but is submitted as a wholly new petition for the species. Please use the following address for all correspondence regarding this petition. -

Jackson Hole Airport Staff Update
JACKSON HOLE AIRPORT STAFF UPDATE April 2020 In This Issue Employee of the Month • Employee of the We are happy to announce that Tony Cross, our Month Human Resources Director, was awarded April’s • Runway Employee of the Month. In Tony’s 5-1/2 years at the Reconstruction airport he has steadfastly put together programs to • Heli-Tours support an enhanced working environment Information and benefits for all employees. He makes a point of • All Teams Meeting being available for anyone needing information, and is committed to working to help us navigate • Jerry Blann benefits or unexpected situations. • Census • Thank You Recently he has worked with various Departments of Public Health related to COVID-19 to make sure that employees had what they needed during their time away from the airport, as well as during their transition back to work. As Jim Elwood stated, “at one point he even drove groceries to someone in Idaho who was not able to get out to the store. He’s just a terrific guy.” Tony, we appreciate your ongoing focus in working with the Board to make Jackson Hole Airport an employer of choice in our area. Congratulations on your selection as Employee of the Month. Latest on the Runway Reconstruction Andrew Wells photo Airport staff, Jviation, the FAA and WYDOT have been working on the runway project since last year. We are excited to share that we have received direction from the FAA and the airport Board, and the runway reconstruction is slated for spring of 2022. The runway project was initially planned as a more simplified rehabilitation or resurfacing, but based on the geotechnical investigation, engineers realized that it will require a full reconstruction. -

Jackson Hole Airport Overview
Air Service Trends at Commercial Airports (WY) Stephen D. Van Beek Select Air Service Committee September 2019 Image Source: Jackson Hole Airport National Aviation Trends in 2019: A Dynamic Industry Sees Accelerating Change What’s In What’s Out Air taxis A-380s Landside Congestion Airside Congestion Metering Roadways and Curbs Providing Free Access 30-year concessions 30-year airline agreements United Airlines Air Transat A220s Bombardier 100/200 Flexible Fixed Non-aeronautical revenues PFCs 2 | September 2019 Air Service Trends at Commercial Airports (WY) Air Service Trends at Commercial Airports 1. Introduction 2. The National Air Service Picture 3. Small Community Air Service: Trends and an Agenda? 4. Air Service: Wyoming Trends 5. Future Air Service Challenges and Policy Ideas to Consider 6. Airport Revenues: Aeronautical and Nonaeronautical 7. An Airport 2020 Agenda 8. Questions and Discussion 3 | September 2019 Air Service Trends at Commercial Airports (WY) The Airport Industry’s SWOT 2019 Strengths Opportunities • Traffic at airport gateways & large hubs (e.g. DEN, DFW) • Passenger growth and low rates providing historic • Airline orderbook of long-haul, fuel efficient, aircraft opportunity for airport capital investments • Liberalization / global traffic diversification • New equity and developer players providing options for • Growing airports’ economic performance development • Dollar easing for some inbound travelers • Technology applications enabling airports to improve the passenger experience and raise new revenues • Low -

Jackson Hole Airport Operating Budget 2019-2020 the Following Document Presents the Jackson Hole Airport Board’S Operating Budget for the Fiscal Year 2019-2020
Jackson Hole Airport Operating Budget 2019-2020 The following document presents the Jackson Hole Airport Board’s operating budget for the Fiscal Year 2019-2020. Jackson Hole Airport Board 1250 E. Airport Rd. Updated 4/11/19 Jackson, WY 83001 307-733-7695 Background The following pages present the Jackson Hole Airport Board’s (the Board’s) operating budget for Fiscal Year 2019/2020. The Town of Jackson and Teton County created the Board by joint resolution in 1967. The Board consists of 5 members jointly appointed by the Town and County. The Joint Powers Agreement sets forth the terms by which the Town, County and Airport operate together. Under this agreement, the Town and County annually review the Airport Budget. The Town and County also both sign all FAA grant agreements as co-sponsors. While the Board operates under the authority of the Town and County, all facilities, equipment, lease holdings and operating rights are owned and managed by the Airport Board. The Board adopted a Certificate of Organization pursuant to the Town of Jackson Ordinance and Board of Teton County Commissioners Resolution on January 2, 1968 officially forming the Airport Board and electing officers. Annually the Certificate of Organization is renewed, and new officers are elected as appointed by the Town and County. For the year February 1, 2019 – January 31, 2020 the slate of officers is: Rick Braun, President; Mary Gibson Scott, Vice President; John Eastman, Treasurer; Bob McLaurin, Secretary; Jerry Blann, Member. The Board operates the Airport inside the boundaries of Grand Teton National Park (the Park) under a Use Agreement with the U.S. -

Annual Privatization Report 2021 — Aviation
ANNUAL PRIVATIZATION REPORT: AVIATION by Robert W. Poole, Jr. Project Director: Austill Stuart July 2021 Reason Foundation’s mission is to advance a free society by developing, applying, and promoting libertarian principles, including individual liberty, free markets, and the rule of law. We use journalism and public policy research to influence the frameworks and actions of policymakers, journalists, and opinion leaders. Reason Foundation’s nonpartisan public policy research promotes choice, competition, and a dynamic market economy as the foundation for human dignity and progress. Reason produces rigorous, peer- reviewed research and directly engages the policy process, seeking strategies that emphasize cooperation, flexibility, local knowledge, and results. Through practical and innovative approaches to complex problems, Reason seeks to change the way people think about issues, and promote policies that allow and encourage individuals and voluntary institutions to flourish. Reason Foundation is a tax-exempt research and education organization as defined under IRS code 501(c)(3). Reason Foundation is supported by voluntary contributions from individuals, foundations, and corporations. The views are those of the author, not necessarily those of Reason Foundation or its trustees. TABLE OF CONTENTS PART 1 INTRODUCTION ................................................................................................................... 1 PART 2 AIRPORTS ............................................................................................................................ -

2013 Sage Grouse Hunting Season Was 10 Days Long, Keeping Opening Day on the 3Rd Saturday in September (Sept
2013 GREATER SAGE-GROUSE JOB COMPLETION REPORT June 1, 2013 – May 31, 2014 Wyoming Game and Fish Department Cheyenne, WY Table of Contents Page Table of Contents ........................................................................................................... i Statewide Summary JCR .............................................................................................. 1 Bates Hole/Shirley Basin JCR ..................................................................................... 61 Big Horn Basin JCR .................................................................................................... 88 Northeast JCR ........................................................................................................... 101 South-Central JCR .................................................................................................... 126 Southwest JCR .......................................................................................................... 147 Upper Green River Basin JCR ................................................................................. 170 Upper Snake River Basin JCR ................................................................................. 196 Wind River/Sweetwater River JCR .......................................................................... 214 Statewide Sage-Grouse Job Completion Report 2013 June 2013-May 2014 Tom Christiansen Wyoming Game & Fish Dept. 1 Wyoming Sage-Grouse Job Completion Report Conservation Plan Area: Statewide Summary Period Covered: 6/1/2013–