(Limpr.) Warnst
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
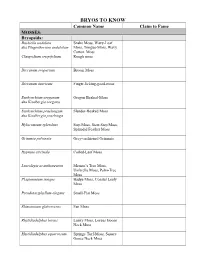
Bryo's to Know Table
BRYOS TO KNOW Common Name Claim to Fame MOSSES: Bryopsida: Buckiella undulata Snake Moss, Wavy-Leaf aka Plagiothecium undulatum Moss, Tongue-Moss, Wavy Cotton, Moss Claopodium crispifolium Rough moss Dicranum scoparium Broom Moss Dicranum tauricum Finger-licking-good-moss Eurhynchium oreganum Oregon Beaked-Moss aka Kindbergia oregana Eurhynchium praelongum Slender-Beaked Moss aka Kindbergia praelonga Hylocomium splendens Step Moss, Stair-Step Moss, Splendid Feather Moss Grimmia pulvinata Grey-cushioned Grimmia Hypnum circinale Coiled-Leaf Moss Leucolepis acanthoneuron Menzie’s Tree Moss, Umbrella Moss, Palm-Tree Moss Plagiomnium insigne Badge Moss, Coastal Leafy Moss Pseudotaxiphyllum elegans Small-Flat Moss Rhizomnium glabrescens Fan Moss Rhytidiadelphus loreus Lanky Moss, Loreus Goose Neck Moss Rhytidiadelphus squarrosum Springy Turf-Moss, Square Goose Neck Moss Rhytidiadelphus triquetrus Electrified Cat-Tail Moss, Goose Necked Moss Rhytidiopsus robusta Robust mountain moss Schistostega pennata Goblin’s Gold, Luminous Moss Polytrichopsida: Atrichum Atrichum Moss , Crane’s Bill Moss (for Atrichum selwynii) Pogonatum contortum Contorted Pogonatum Moss Polytrichum commune Common Hair Cap Moss Polytrichum piliferum Bristly Haircap Moss Andreaeopsida Andreaea nivalis Granite moss, Lantern moss, Snow Rock Moss Sphagnopsida: Sphagnum capillifolium Red Bog Moss, Small Red Peat Moss Sphagnum papillosum Fat Bog Moss, Papillose sphagnum Sphagnum squarrosum Shaggy Sphagnum, Spread- Leaved Peat Moss Takakiopsida: Takakia lepidoziooides Impossible -

Coptis Trifolia Conservation Assessment
CONSERVATION ASSESSMENT for Coptis trifolia (L.) Salisb. Originally issued as Management Recommendations December 1998 Marty Stein Reconfigured-January 2005 Tracy L. Fuentes USDA Forest Service Region 6 and USDI Bureau of Land Management, Oregon and Washington CONSERVATION ASSESSMENT FOR COPTIS TRIFOLIA Table of Contents Page List of Tables ................................................................................................................................. 2 List of Figures ................................................................................................................................ 2 Summary........................................................................................................................................ 4 I. NATURAL HISTORY............................................................................................................. 6 A. Taxonomy and Nomenclature.......................................................................................... 6 B. Species Description ........................................................................................................... 6 1. Morphology ................................................................................................................... 6 2. Reproductive Biology.................................................................................................... 7 3. Ecological Roles ............................................................................................................. 7 C. Range and Sites -

Floristic Study of Bryophytes in a Subtropical Forest of Nabeup-Ri at Aewol Gotjawal, Jejudo Island
− pISSN 1225-8318 Korean J. Pl. Taxon. 48(1): 100 108 (2018) eISSN 2466-1546 https://doi.org/10.11110/kjpt.2018.48.1.100 Korean Journal of ORIGINAL ARTICLE Plant Taxonomy Floristic study of bryophytes in a subtropical forest of Nabeup-ri at Aewol Gotjawal, Jejudo Island Eun-Young YIM* and Hwa-Ja HYUN Warm Temperate and Subtropical Forest Research Center, National Institute of Forest Science, Seogwipo 63582, Korea (Received 24 February 2018; Revised 26 March 2018; Accepted 29 March 2018) ABSTRACT: This study presents a survey of bryophytes in a subtropical forest of Nabeup-ri, known as Geumsan Park, located at Aewol Gotjawal in the northwestern part of Jejudo Island, Korea. A total of 63 taxa belonging to Bryophyta (22 families 37 genera 44 species), Marchantiophyta (7 families 11 genera 18 species), and Antho- cerotophyta (1 family 1 genus 1 species) were determined, and the liverwort index was 30.2%. The predominant life form was the mat form. The rates of bryophytes dominating in mesic to hygric sites were higher than the bryophytes mainly observed in xeric habitats. These values indicate that such forests are widespread in this study area. Moreover, the rock was the substrate type, which plays a major role in providing micro-habitats for bryophytes. We suggest that more detailed studies of the bryophyte flora should be conducted on a regional scale to provide basic data for selecting indicator species of Gotjawal and evergreen broad-leaved forests on Jejudo Island. Keywords: bryophyte, Aewol Gotjawal, liverwort index, life-form Jejudo Island was formed by volcanic activities and has geological, ecological, and cultural aspects (Jeong et al., 2013; unique topological and geological features. -

Riparian Bryophytes of the H. J. Andrews Experimental Forest in the Western Cascades, Oregon
The Bryologist 99(2), pp. 226-235 Copyright © 1996 by the American Bryological and Lichenological Society, Inc. Riparian Bryophytes of the H. J. Andrews Experimental Forest in the Western Cascades, Oregon BENGT GUNNAR JONSSON I Department of Forest Science, Oregon State University, Corvallis, OR 97331 Abstract. The knowledge of the distribution and habitat demands for bryophytes in the Pacific Northwest is scarce, and few published quantitative accounts of the flora are present. The present paper includes habitat description, elevational range, substrate preference, and frequency esti- mates for more than 130 riparian mosses and liverworts found in old-growth Pseudotsuga-Tsuga forests of the H. J. Andrews Experimental Forest, Oregon. The data are based on 360 samples distributed among 42 sites covering 1st to 5th order streams and 420 to 1250 m. TWINSPAN analysis resulted in 6 sample groups, representing samples from different elevations, geomorphic surfaces, and stream sizes. The most common mosses are Eurhynchium oreganum, Isothecium stoloniferum, Hypnum circinale, and Dicranum fuscescens. Among the hepatics Scapania bolan- deri, Cephalozia lunulifolia, and Porella navicularis are the most abundant species. Most species are rare at both site and sampte gtot (ever; and this is especially true for acrocarps where more than one-third of the observed species occurred in only one or two sites orland samples. Four of the occurring species (i.e., Antitrichia curtipendula, Buxbaumia piperi, Douinia ovata, and Ptili- dium californicum) are listed for special management and/or regional surveys. Bryophytes constitute an important and conspic- a wide range of different substrates, disturbance uous component of old-growth forests in the Pacific patterns, and a moist microclimate are important Northwest (Lesica et al. -

Old Woman Creek National Estuarine Research Reserve Management Plan 2011-2016
Old Woman Creek National Estuarine Research Reserve Management Plan 2011-2016 April 1981 Revised, May 1982 2nd revision, April 1983 3rd revision, December 1999 4th revision, May 2011 Prepared for U.S. Department of Commerce Ohio Department of Natural Resources National Oceanic and Atmospheric Administration Division of Wildlife Office of Ocean and Coastal Resource Management 2045 Morse Road, Bldg. G Estuarine Reserves Division Columbus, Ohio 1305 East West Highway 43229-6693 Silver Spring, MD 20910 This management plan has been developed in accordance with NOAA regulations, including all provisions for public involvement. It is consistent with the congressional intent of Section 315 of the Coastal Zone Management Act of 1972, as amended, and the provisions of the Ohio Coastal Management Program. OWC NERR Management Plan, 2011 - 2016 Acknowledgements This management plan was prepared by the staff and Advisory Council of the Old Woman Creek National Estuarine Research Reserve (OWC NERR), in collaboration with the Ohio Department of Natural Resources-Division of Wildlife. Participants in the planning process included: Manager, Frank Lopez; Research Coordinator, Dr. David Klarer; Coastal Training Program Coordinator, Heather Elmer; Education Coordinator, Ann Keefe; Education Specialist Phoebe Van Zoest; and Office Assistant, Gloria Pasterak. Other Reserve staff including Dick Boyer and Marje Bernhardt contributed their expertise to numerous planning meetings. The Reserve is grateful for the input and recommendations provided by members of the Old Woman Creek NERR Advisory Council. The Reserve is appreciative of the review, guidance, and council of Division of Wildlife Executive Administrator Dave Scott and the mapping expertise of Keith Lott and the late Steve Barry. -

STUTTGARTER BEITRÄGE ZUR NATURKUNDE Ser
download Biodiversity Heritage Library, http://www.biodiversitylibrary.org/ Stuttgarter Beitrage zur Naturkunde 3V< .... f Serie A (Biologie) 1^ Vt" Herausgeber: Staatliches Museum für Naturkunde, Rosenstein 1, D-70191 Stuttgart Stuttgarter Beitr. Naturk. Ser. A Nr. 589 15 S. Stuttgart, 10. 8. 1999 New Views on the Relationships among European Pleurocarpous Mosses By Lars Hedenäs, Stockholm With 2 figures Summary An overview is given of the results of higher level cladistic studies of pleurocarpous moss- es, and the implications of these for the classification of severel larger families represented in Europe. Starting with the more ancestral taxa, the traditional Isobryales forms a basal grade, followed by another grade including taxa with capsules of Brachytbecwm-sha.pc. The latter grade includes taxa such as the Brachytheciaceae, Ctenidiaceae, and Hylocomiaceae, as well as the subfamily Heterocladioideae of the Thuidiaceae. The Amblystegiaceae, Rhytidiaceae, the temperate members of the Hypnaceae, and the Thuidiceae (excl. the Heterocladioideae) form a monophyletic group, with the Plagiotheciaecae as their sister group. The few European members of the Callicostaceae, Hokeriaceae, Leucomiaceae, and Sematophyllaceae belong to another monophyletic group with mainly tropical and subtropical members. The tropical members of the traditionally heterogeneous Hypnaceae are not closely related to the temper- ate members found in Europe. Zusammenfassung Die Arbeit stellt die Ergebnisse einer Stammbaumanalyse der Großgruppen pleurokarper Moose und ihre Auswirkungen auf die Einteilung einiger größerer in Europa vertretener Fa- milien dar. Wenn man mit den ursprünglichen Gruppen beginnt, stehen die Isobryales im her- kömmlichen Sinn an der Basis, darauf folgen als weitere Verwandschaftsgruppe die Vertreter mit Brachythecwm-artig gebauten Kapseln. Letztere umfaßt Gruppen wie die Brachythecia- ceae, Ctenidiaceae und Hylocomiaceae sowie die Unterfamilie Heterocladioideae der Thui- diaceae. -

Keys for the Determination of Families of Pleurocarpous Mosses of Africa
Keys for the determination of families of pleurocarpous mosses of Africa E. Petit Extracted from: Cléfs pour la determination des familles et des genres des mousses pleurocarpes (Musci) d'AfriqueBull. Jard. Bot. Nat. Belg. 48: 135-181 (1978) Translated by M.J.Wigginton, 36 Big Green, Warmington, Peterborough, PE and C.R. Stevenson, 111 Wootton Road, King's Lynn, Norfolk, PE The identification of tropical African mosses is fraught with difficulty, not least because of the sparseness of recent taxonomic literature. Even the determination of specimens to family or genus can be problematical. The paper by Petit (1978) is a valiant attempt to provide workable keys (and short descriptions) to all the families and genera of African pleurocarpous mosses, and remains the only such comprehensive treatment. Whilst the shortcomings of any such keys apply, the keys have nonetheless proved to be of assistance in placing specimens in taxonomic groups. However, for the non-French reader, the use of the keys can be a tedious business, necessitating frequent recourse to dictionaries and grammars. Members of the BBS have made collections in a number of tropical African countries in recent years, including on the BBS expedition to Malawi and privately to Madagascar, Tanzania and Zaire. This provided the impetus for making a translation of Petit's keys. Neither of us is an expert linguist, and doubtless in places, some of the subtleties of the language have escaped us. A rather free translation has sometimes proved necessary in order to give the sense of the text. Magill's Glossarium Polyglottum Bryologicae has been valuable in assisting with technical terms. -

Flora of New Zealand Mosses
FLORA OF NEW ZEALAND MOSSES BRACHYTHECIACEAE A.J. FIFE Fascicle 46 – JUNE 2020 © Landcare Research New Zealand Limited 2020. Unless indicated otherwise for specific items, this copyright work is licensed under the Creative Commons Attribution 4.0 International licence Attribution if redistributing to the public without adaptation: "Source: Manaaki Whenua – Landcare Research" Attribution if making an adaptation or derivative work: "Sourced from Manaaki Whenua – Landcare Research" See Image Information for copyright and licence details for images. CATALOGUING IN PUBLICATION Fife, Allan J. (Allan James), 1951- Flora of New Zealand : mosses. Fascicle 46, Brachytheciaceae / Allan J. Fife. -- Lincoln, N.Z. : Manaaki Whenua Press, 2020. 1 online resource ISBN 978-0-947525-65-1 (pdf) ISBN 978-0-478-34747-0 (set) 1. Mosses -- New Zealand -- Identification. I. Title. II. Manaaki Whenua-Landcare Research New Zealand Ltd. UDC 582.345.16(931) DC 588.20993 DOI: 10.7931/w15y-gz43 This work should be cited as: Fife, A.J. 2020: Brachytheciaceae. In: Smissen, R.; Wilton, A.D. Flora of New Zealand – Mosses. Fascicle 46. Manaaki Whenua Press, Lincoln. http://dx.doi.org/10.7931/w15y-gz43 Date submitted: 9 May 2019 ; Date accepted: 15 Aug 2019 Cover image: Eurhynchium asperipes, habit with capsule, moist. Drawn by Rebecca Wagstaff from A.J. Fife 6828, CHR 449024. Contents Introduction..............................................................................................................................................1 Typification...............................................................................................................................................1 -

Molecular Phylogeny of Chinese Thuidiaceae with Emphasis on Thuidium and Pelekium
Molecular Phylogeny of Chinese Thuidiaceae with emphasis on Thuidium and Pelekium QI-YING, CAI1, 2, BI-CAI, GUAN2, GANG, GE2, YAN-MING, FANG 1 1 College of Biology and the Environment, Nanjing Forestry University, Nanjing 210037, China. 2 College of Life Science, Nanchang University, 330031 Nanchang, China. E-mail: [email protected] Abstract We present molecular phylogenetic investigation of Thuidiaceae, especially on Thudium and Pelekium. Three chloroplast sequences (trnL-F, rps4, and atpB-rbcL) and one nuclear sequence (ITS) were analyzed. Data partitions were analyzed separately and in combination by employing MP (maximum parsimony) and Bayesian methods. The influence of data conflict in combined analyses was further explored by two methods: the incongruence length difference (ILD) test and the partition addition bootstrap alteration approach (PABA). Based on the results, ITS 1& 2 had crucial effect in phylogenetic reconstruction in this study, and more chloroplast sequences should be combinated into the analyses since their stability for reconstructing within genus of pleurocarpous mosses. We supported that Helodiaceae including Actinothuidium, Bryochenea, and Helodium still attributed to Thuidiaceae, and the monophyletic Thuidiaceae s. lat. should also include several genera (or species) from Leskeaceae such as Haplocladium and Leskea. In the Thuidiaceae, Thuidium and Pelekium were resolved as two monophyletic groups separately. The results from molecular phylogeny were supported by the crucial morphological characters in Thuidiaceae s. lat., Thuidium and Pelekium. Key words: Thuidiaceae, Thuidium, Pelekium, molecular phylogeny, cpDNA, ITS, PABA approach Introduction Pleurocarpous mosses consist of around 5000 species that are defined by the presence of lateral perichaetia along the gametophyte stems. Monophyletic pleurocarpous mosses were resolved as three orders: Ptychomniales, Hypnales, and Hookeriales (Shaw et al. -

Genetic Variation in the Clonal Bryophyte Hylocomium Splendens at Hierarchical Geographical Scales in Scandinavia
Heredity 78 (1997) 293-301 Received 26 April 1996 Genetic variation in the clonal bryophyte Hylocomium splendens at hierarchical geographical scales in Scandinavia NILS CRONBERG*t, ULF MOLAU:j: & MATS SONESSON§ tDepartment of Systematic Botany, University of Lund, 6. Val/gatan 18-20, S-223 61 Lund, tDepartment of Systematic Botany, University of Goteborg, Carl Skottsbergs Gata 22, S-413 19 Goteborg and §Abisko Scientific Research Station, The Royal Swedish Academy of Sciences, S-981 07 Abisko, Sweden The widespread bryophyte Hylocomium splendens was sampled in a hierarchical fashion from populations representing four Scandinavian vegetation zones. Allozyme electrophoresis revealed variation at 11 out of 13 screened loci, allowing accurate identification of genotypes. From a total sample of 298 shoots 79 genotypes could be detected, giving the proportion of distinguishable genotypes (PD) of 0.265. The total allelic diversity (HT) based on polymorphic loci was 0.274. The relative differentiation among populations was low (GsT = 0.073), indicating a high level of gene flow. Differences in population structuring occurred between a subarctic alpine site vs. three lowland sites. The subarctic-alpine population had one widespread clone, which appeared to be propagated by dispersal of vegetative fragments. That population also comprised many rare genotypes, often occurring together within 10 x 10 cm patches. The lowland populations had genotypes that were locally common and often dominant within the patches. When identical genotypes were observed in multiple patches within these populations, it was usually statistically highly probable that they had arisen by independent sexual recombinations. Keywords: allozyme, Bryophyta, clonal plant, genetic diversity, hierarchical variation, Hy/oco mium splendens. -

About the Book the Format Acknowledgments
About the Book For more than ten years I have been working on a book on bryophyte ecology and was joined by Heinjo During, who has been very helpful in critiquing multiple versions of the chapters. But as the book progressed, the field of bryophyte ecology progressed faster. No chapter ever seemed to stay finished, hence the decision to publish online. Furthermore, rather than being a textbook, it is evolving into an encyclopedia that would be at least three volumes. Having reached the age when I could retire whenever I wanted to, I no longer needed be so concerned with the publish or perish paradigm. In keeping with the sharing nature of bryologists, and the need to educate the non-bryologists about the nature and role of bryophytes in the ecosystem, it seemed my personal goals could best be accomplished by publishing online. This has several advantages for me. I can choose the format I want, I can include lots of color images, and I can post chapters or parts of chapters as I complete them and update later if I find it important. Throughout the book I have posed questions. I have even attempt to offer hypotheses for many of these. It is my hope that these questions and hypotheses will inspire students of all ages to attempt to answer these. Some are simple and could even be done by elementary school children. Others are suitable for undergraduate projects. And some will take lifelong work or a large team of researchers around the world. Have fun with them! The Format The decision to publish Bryophyte Ecology as an ebook occurred after I had a publisher, and I am sure I have not thought of all the complexities of publishing as I complete things, rather than in the order of the planned organization. -

Hypnaceaeandpossiblyrelatedfn
Hikobial3:645-665.2002 Molecularphylo窪enyOfhypnobrJ/aleanmOssesasin化rredfroma lar淫e-scaledatasetofchlOroplastlbcL,withspecialre他rencetothe HypnaceaeandpOssiblyrelatedfnmilies1 HIRoMITsuBoTA,ToMoTsuGuARIKAwA,HIRoYuKIAKIYAMA,EFRAINDELuNA,DoLoREs GoNzALEz,MASANoBuHIGucHIANDHIRoNoRIDEGucHI TsuBoTA,H、,ARIKAwA,T,AKIYAMA,H,,DELuNA,E,GoNzALEz,,.,HIGucHI,M 4 &DEGucHI,H、2002.Molecularphylogenyofhypnobryaleanmossesasinferred fiPomalarge-scaledatasetofchloroplastr6cL,withspecialreferencetotheHypnaceae andpossiblyrelatedfamiliesl3:645-665. ▲ Phylogeneticrelationshipswithinthehypnobryaleanmosses(ie,theHypnales,Leuco- dontales,andHookeriales)havebeenthefbcusofmuchattentioninrecentyears Herewepresentphylogeneticinfierencesonthislargeclade,andespeciallyonthe Hypnaceaeandpossiblyrelatedftlmilies,basedonmaximumlikelihoodanalysisof l81r6cLsequences、Oursmdycorroboratesthat(1)theHypnales(sstr.[=sensu Vittl984])andLeucodontalesareeachnotmonophyleticentities、TheHypnalesand LeucodontalestogethercompriseawellsupportedsistercladetotheHookeriales;(2) theSematophyllaceae(s」at[=sensuTsubotaetaL2000,2001a,b])andPlagiothecia‐ ceae(s・str.[=sensupresentDareeachresolvedasmonophyleticgroups,whileno particularcladeaccommodatesallmembersoftheHypnaceaeandCryphaeaceae;and (3)theHypnaceaeaswellasitstypegenusノリDlwz"川tselfwerepolyphyletioThese resultsdonotconcurwiththesystemsofVitt(1984)andBuckandVitt(1986),who suggestedthatthegroupswithasinglecostawouldhavedivergedfiFomthehypnalean ancestoratanearlyevolutionarystage,fbllowedbythegroupswithadoublecosta (seealsoTsubotaetall999;Bucketal2000)OurresultsfiPomlikelihoodanalyses