Molecular Identification and Distribution of Native and Exotic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Taxonomic Assessment of Lumbricidae (Oligochaeta) Earthworm Genera Using DNA Barcodes
European Journal of Soil Biology 48 (2012) 41e47 Contents lists available at SciVerse ScienceDirect European Journal of Soil Biology journal homepage: http://www.elsevier.com/locate/ejsobi Original article Taxonomic assessment of Lumbricidae (Oligochaeta) earthworm genera using DNA barcodes Marcos Pérez-Losada a,*, Rebecca Bloch b, Jesse W. Breinholt c, Markus Pfenninger b, Jorge Domínguez d a CIBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos, Universidade do Porto, Campus Agrário de Vairão, 4485-661 Vairão, Portugal b Biodiversity and Climate Research Centre, Lab Centre, Biocampus Siesmayerstraße, 60323 Frankfurt am Main, Germany c Department of Biology, Brigham Young University, Provo, UT 84602-5181, USA d Departamento de Ecoloxía e Bioloxía Animal, Universidade de Vigo, E-36310, Spain article info abstract Article history: The family Lumbricidae accounts for the most abundant earthworms in grasslands and agricultural Received 26 May 2011 ecosystems in the Paleartic region. Therefore, they are commonly used as model organisms in studies of Received in revised form soil ecology, biodiversity, biogeography, evolution, conservation, soil contamination and ecotoxicology. 14 October 2011 Despite their biological and economic importance, the taxonomic status and evolutionary relationships Accepted 14 October 2011 of several Lumbricidae genera are still under discussion. Previous studies have shown that cytochrome c Available online 30 October 2011 Handling editor: Stefan Schrader oxidase I (COI) barcode phylogenies are informative at the intrageneric level. Here we generated 19 new COI barcodes for selected Aporrectodea specimens in Pérez-Losada et al. [1] including nine species and 17 Keywords: populations, and combined them with all the COI sequences available in Genbank and Briones et al. -

Study on the Biomass and Productivity of Lumbricidae Populations in the Deciduous Ecosystem
Muzeul Olteniei Craiova. Oltenia. Studii úi comunicări. ùtiinĠele Naturii. Tom. 28, No. 2/2012 ISSN 1454-6914 STUDY ON THE BIOMASS AND PRODUCTIVITY OF LUMBRICIDAE POPULATIONS IN THE DECIDUOUS ECOSYSTEM BRÎNZEA GheorghiĠa Abstract. The present study analyses the earthworms in the deciduous ecosystem (Ruginoasa Station) of Cândeúti Platform, in the south-east of Arge܈ County, in terms of biomass and monthly productivity during March-October 2007. Besides numerical dominance, the biomass of Lumbricidae species highlights the role of each species in the activity of matter and energy transfer within an ecosystem. The special dynamics of the two parameters showed different values, given the structure of the vegetation, soil and soil layers, and especially the differences between the individual weights of the species. Octolasion lacteum species was dominant in terms of biomass density and recorded high values at the soil levels analysed, except in the litter. Of the total calculated biomass (27.598 mg d.s./square meter), this species dominates in a ratio of 74.69%. Biological productivity was increased at 0-10 cm, with the highest value for Octolasion lacteum species (4.03 mg d.s./m2), followed by Eisenia lucens species (0.9 mg d.s./m2) and Aporrectodea rosea rosea species (0.641 mg d.s./m2 ). Also, the highest values of the biomass were recorded in spring and autumn months. The species with a lower numerical density recorded low values of the biomass, leading to low biological productivity and vice versa. Keywords: deciduous forest, lumbricidae, biomass, productivity. Rezumat. Studiu privind biomasa úi productivitatea populaĠiilor de Lumbricidae într-un ecosistem de foioase. -

Impact of Agricultural Practices on Biodiversity of Soil Invertebrates
Impact of Agricultural Practices on Biodiversity of Soil Invertebrates Impact of • Stefano Bocchi and Francesca Orlando Agricultural Practices on Biodiversity of Soil Invertebrates Edited by Stefano Bocchi and Francesca Orlando Printed Edition of the Special Issue Published in Agronomy www.mdpi.com/journal/agronomy Impact of Agricultural Practices on Biodiversity of Soil Invertebrates Impact of Agricultural Practices on Biodiversity of Soil Invertebrates Editors Stefano Bocchi Francesca Orlando MDPI • Basel • Beijing • Wuhan • Barcelona • Belgrade • Manchester • Tokyo • Cluj • Tianjin Editors Stefano Bocchi Francesca Orlando University of Milan University of Milan Italy Italy Editorial Office MDPI St. Alban-Anlage 66 4052 Basel, Switzerland This is a reprint of articles from the Special Issue published online in the open access journal Agronomy (ISSN 2073-4395) (available at: https://www.mdpi.com/journal/agronomy/special issues/Soil Invertebrates). For citation purposes, cite each article independently as indicated on the article page online and as indicated below: LastName, A.A.; LastName, B.B.; LastName, C.C. Article Title. Journal Name Year, Volume Number, Page Range. ISBN 978-3-03943-719-1 (Hbk) ISBN 978-3-03943-720-7 (PDF) Cover image courtesy of Valentina Vaglia. c 2020 by the authors. Articles in this book are Open Access and distributed under the Creative Commons Attribution (CC BY) license, which allows users to download, copy and build upon published articles, as long as the author and publisher are properly credited, which ensures maximum dissemination and a wider impact of our publications. The book as a whole is distributed by MDPI under the terms and conditions of the Creative Commons license CC BY-NC-ND. -
Size Variation and Geographical Distribution of the Luminous Earthworm Pontodrilus Litoralis (Grube, 1855) (Clitellata, Megascolecidae) in Southeast Asia and Japan
A peer-reviewed open-access journal ZooKeys 862: 23–43 (2019) Size variation and distribution of Pontodrilus litoralis 23 doi: 10.3897/zookeys.862.35727 RESEARCH ARTICLE http://zookeys.pensoft.net Launched to accelerate biodiversity research Size variation and geographical distribution of the luminous earthworm Pontodrilus litoralis (Grube, 1855) (Clitellata, Megascolecidae) in Southeast Asia and Japan Teerapong Seesamut1,2,4, Parin Jirapatrasilp2, Ratmanee Chanabun3, Yuichi Oba4, Somsak Panha2 1 Biological Sciences Program, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand 2 Ani- mal Systematics Research Unit, Department of Biology, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand 3 Program in Animal Science, Faculty of Agriculture Technology, Sakon Nakhon Rajabhat University, Sakon Nakhon 47000, Thailand 4 Department of Environmental Biology, Chubu University, Kasugai 487-8501, Japan Corresponding authors: Somsak Panha ([email protected]), Yuichi Oba ([email protected]) Academic editor: Samuel James | Received 24 April 2019 | Accepted 13 June 2019 | Published 9 July 2019 http://zoobank.org/663444CA-70E2-4533-895A-BF0698461CDF Citation: Seesamut T, Jirapatrasilp P, Chanabun R, Oba Y, Panha S (2019) Size variation and geographical distribution of the luminous earthworm Pontodrilus litoralis (Grube, 1855) (Clitellata, Megascolecidae) in Southeast Asia and Japan. ZooKeys 862: 23–42. https://doi.org/10.3897/zookeys.862.35727 Abstract The luminous earthworm Pontodrilus litoralis (Grube, 1855) occurs in a very wide range of subtropical and tropical coastal areas. Morphometrics on size variation (number of segments, body length and diameter) and genetic analysis using the mitochondrial cytochrome c oxidase subunit 1 (COI) gene sequence were conducted on 14 populations of P. -
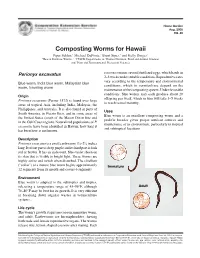
Composting Worms for Hawaii
Home Garden Aug. 2005 HG-46 Composting Worms for Hawaii Piper Selden,1 Michael DuPonte,2 Brent Sipes,3 and Kelly Dinges2 1Hawaii Rainbow Worms, 2, 3CTAHR Departments of 2Human Nutrition, Food and Animal Sciences and 3Plant and Environmental Protection Sciences Perionyx excavatus cocoon contains several fertilized eggs, which hatch in 2–3 weeks under suitable conditions. Reproductive rates Blue worm, India blue worm, Malaysian blue vary according to the temperature and environmental worm, traveling worm conditions, which in vermiculture depend on the maintenance of the composting system. Under favorable Origin conditions, blue worms may each produce about 20 offspring per week, which in turn will take 3–5 weeks Perionyx excavatus (Perrier 1872) is found over large to reach sexual maturity. areas of tropical Asia, including India, Malaysia, the Phillippines, and Australia. It is also found in parts of Uses South America, in Puerto Rico, and in some areas of Blue worm is an excellent composting worm and a the United States (south of the Mason Dixon line and prolific breeder given proper nutrient sources and in the Gulf Coast region). Naturalized populations of P. maintenance of its environment, particularly in tropical excavatus have been identified in Hawaii; how long it and subtropical locations. has been here is not known. Description 1 3 Perionyx excavatus is a small earthworm 1 ⁄4–2 ⁄4 inches long. Its front part is deep purple and its hind part is dark red or brown. It has an iridescent, blue-violet sheen on its skin that is visible in bright light. These worms are highly active and twitch when disturbed. -

Phylogenetic and Phenetic Systematics of The
195 PHYLOGENETICAND PHENETICSYSTEMATICS OF THE OPISTHOP0ROUSOLIGOCHAETA (ANNELIDA: CLITELLATA) B.G.M. Janieson Departnent of Zoology University of Queensland Brisbane, Australia 4067 Received September20, L977 ABSTMCT: The nethods of Hennig for deducing phylogeny have been adapted for computer and a phylogran has been constructed together with a stereo- phylogran utilizing principle coordinates, for alL farnilies of opisthopor- ous oligochaetes, that is, the Oligochaeta with the exception of the Lunbriculida and Tubificina. A phenogran based on the sane attributes conpares unfavourably with the phyLogralnsin establishing an acceptable classification., Hennigrs principle that sister-groups be given equal rank has not been followed for every group to avoid elevation of the more plesionorph, basal cLades to inacceptabl.y high ranks, the 0ligochaeta being retained as a Subclass of the class Clitellata. Three orders are recognized: the LumbricuLida and Tubificida, which were not conputed and the affinities of which require further investigation, and the Haplotaxida, computed. The Order Haplotaxida corresponds preciseLy with the Suborder Opisthopora of Michaelsen or the Sectio Diplotesticulata of Yanaguchi. Four suborders of the Haplotaxida are recognized, the Haplotaxina, Alluroidina, Monil.igastrina and Lunbricina. The Haplotaxina and Monili- gastrina retain each a single superfanily and fanily. The Alluroidina contains the superfamiJ.y All"uroidoidea with the fanilies Alluroididae and Syngenodrilidae. The Lurnbricina consists of five superfaniLies. -

The Giant Palouse Earthworm (Driloleirus Americanus)
PETITION TO LIST The Giant Palouse Earthworm (Driloleirus americanus) AS A THREATENED OR ENDANGERED SPECIES UNDER THE ENDANGERED SPECIES ACT June 30, 2009 Friends of the Clearwater Center for Biological Diversity Palouse Audubon Palouse Prairie Foundation Palouse Group of the Sierra Club 1 June 30, 2009 Ken Salazar, Secretary of the Interior Robyn Thorson, Regional Director U.S. Department of the Interior U.S. Fish & Wildlife Service 1849 C Street N.W. Pacific Region Washington, DC 20240 911 NE 11th Ave Portland, Oregon Dear Secretary Salazar, Friends of the Clearwater, Center for Biological Diversity, Palouse Prairie Foundation, Palouse Audubon, Palouse Group of the Sierra Club and Steve Paulson formally petition to list the Giant Palouse Earthworm (Driloleirus americanus) as a threatened or endangered species pursuant to the Endangered Species Act (”ESA”), 16 U.S.C. §1531 et seq. This petition is filed under 5 U.S.C. 553(e) and 50 CFR 424.14 (1990), which grant interested parties the right to petition for issuance of a rule from the Secretary of Interior. Petitioners also request that critical habitat be designated for the Giant Palouse Earthworm concurrent with the listing, pursuant to 50 CFR 424.12, and pursuant to the Administrative Procedures Act (5 U.S.C. 553). The Giant Palouse Earthworm (D. americanus) is found only in the Columbia River Drainages of eastern Washington and Northern Idaho. Only four positive collections of this species have been made within the last 110 years, despite the fact that the earthworm was historically considered “very abundant” (Smith 1897). The four collections include one between Moscow, Idaho and Pullman, Washington, one near Moscow Mountain, Idaho, one at a prairie remnant called Smoot Hill and a fourth specimen near Ellensberg, Washington (Fender and McKey- Fender, 1990, James 2000, Sánchez de León and Johnson-Maynard, 2008). -

Page 1-21.FH10
ISSN 0375-1511 Rec. zool. Surv. India: 113(Part-4): 213-227,2013 REPORT ON THE SOIL FAUNA OF BHADRAK AND BALASORE DISTRICT, ORISSA RiNKU GoswAMi, MAYA GHOSH AND DEBDULAL SAHA Zoological Survey of India, M-Block, New Alipore, Kolkata-700053 INTRODUCTION In this study, the assessment of soil fauna in Soil is one of the basic natural resoiirces that the study areas aimed at obtaining a general supports life on Earth. It is a huge ecosystem, overview of soil fauna in the ecosystems of the which is the habitat to several living organisms. region. Perusal of published literature shows no Historically, most of the efforts on biodiversity such systematic study was conducted in these studies focused, especially on aboveground plant areas of our study zone previously. and animal species (Wardle, 2006). However, it is Soil Fauna and their Function in Soil well recognized that in most terrestrial There are many animal groups inhabiting soil ecosystems, the belowground biota supports system. It has been reported that of the total much greater diversity of organisms than does the nirmber of described species on Earth (~1,500,000), aboveground biota, because soils are the central as many as 23 per cent are soil animals (Decaens et. organising entities in terrestrial ecosystems al., 2006). Estimated nirmbers of soil species include (Coleman, and Whitman, 2005). Soil fauna is a 30,000 bacteria; 1,500,000 fungi; 60,000 algae; 10,000 highly diverse group of organisms living within protozoa; 500,000 nematodes; and 3,000 the soil and make soil alive by their activity. -

Checklist of New Zealand Earthworms Updated from Lee (1959) by R.J
Checklist of New Zealand Earthworms updated from Lee (1959) by R.J. Blakemore August, 2006 COE fellow, Soil Ecology Group, Yokohama National Univeristy, Japan. Summary This review is based on Blakemore (2004) that updated the work completed over 40 years earlier by Lee (1959) as modified by Blakemore in Lee et al. , (2000) and in Glasby et al. (2007/8) based on the information presented at the "Species 2000" meeting held Jan. 2000 at Te Papa Museum in Wellington, New Zealand. In the current checklist Acanthodrilidae, Octochaetidae and Megascolecidae sensu Blakemore (2000b) are all given separate family status. Whereas Lee (1959) listed approximately 193 species, the current list has about 199 taxa. Because many of the natives have few reports, or are based on only a few specimens, approximately 77 are listed as "threatened" or "endangered" in Dept. Conservation threatened species list (see www.doc.govt.nz/Conservation/001~Plants-and-Animals/006~Threatened-species/Terrestrial-invertebrate-(part-one).asp April, 2005) and three species are detailed in McGuinness (2001). Further studies such as those of Springett & Grey (1998) are required. Currently I seek funding to complete my database into an interactive guide to species, to use to conduct surveys in New Zealand. Some of the changes in Blakemore (2004) from Lee (1959) are: • Microscolex macquariensis (Beddard, 1896) is removed from the list because it is known only from Macquarie Island, which is now claimed as Australian territory (see Blakemore, 2000b). • Megascolides orthostichon (Schmarda, 1861) is removed from the fauna as Fletcher (1886: 524) reported that “on the authority of Captain Hutton” this species was not from New Zealand and may be from Mt Wellington in Tasmania (see Blakemore, 2000b). -

Asian Jumping Worm (Megascolecidae) Impacts on Physical and Biological Characteristics of Turfgrass Ecosystems
Colby College Digital Commons @ Colby Honors Theses Student Research 2019 Asian Jumping Worm (Megascolecidae) Impacts on Physical and Biological Characteristics of Turfgrass Ecosystems Ella L. Maddi Colby College Follow this and additional works at: https://digitalcommons.colby.edu/honorstheses Part of the Environmental Sciences Commons, and the Soil Science Commons Colby College theses are protected by copyright. They may be viewed or downloaded from this site for the purposes of research and scholarship. Reproduction or distribution for commercial purposes is prohibited without written permission of the author. Recommended Citation Maddi, Ella L., "Asian Jumping Worm (Megascolecidae) Impacts on Physical and Biological Characteristics of Turfgrass Ecosystems" (2019). Honors Theses. Paper 965. https://digitalcommons.colby.edu/honorstheses/965 This Honors Thesis (Open Access) is brought to you for free and open access by the Student Research at Digital Commons @ Colby. It has been accepted for inclusion in Honors Theses by an authorized administrator of Digital Commons @ Colby. Asian Jumping Worm impacts (Megascolecidae) on Physical and Biological Characteristics of Turfgrass Ecosystems An Honors Thesis presented to the Faculty of the Department of Biology at Colby College in partial fulfillment of the requirements for the Degree of Bachelor of Arts with Honors By Ella Maddi Waterville, ME May 20, 2019 Asian Jumping Worm impacts (Megascolecidae) on Physical and Biological Characteristics of Turfgrass Ecosystems An Honors Thesis presented -

Peregrine Earthworms (Clitellata: Lumbricidae) from Bulgaria and Turkey
Annuaire de l’Université de Sofia“ St. Kliment Ohridski” Faculte de Biologie 2017, volume 102, livre 4, pp. 45-53 Youth Scientific Conference “Kliment’s Days”, Sofia 2016 PEREGRINE EARTHWORMS (CLITELLATA: LUMBRICIDAE) FROM BULGARIA AND TURKEY METE MISIRLIOĞLU1, HRISTO VALCHOVSKI2* , RALITSA TSEKOVA3 1-Eskişehir Osmangazi University, Faculty of Science and Letters, Department of Biology, 26480 Eskişehir, Turkey. 2-Institute of Soil Science, Agrotechnologies and Plant Protection “N. Poushkarov”, 7 Shosse Bankya Str., 1080 Sofia, Bulgaria. 3-Sofia University, Faculty of Biology, Department of Ecology, 8 Dragan Tzankov blvd., 1164 Sofia, Bulgaria * Corresponding author: [email protected] Keywords: Earthworms, Lumbricidae, Peregrine, Bulgaria, Turkey Abstract: In this paper we summarize the knowledge on peregrine earthworm fauna from the entire area of Bulgaria and Turkey. The peregrine lumbricids from Bulgaria and Turkey contains 16 taxa belonging to 7 genera. The most common peregrine taxa of Bulgaria are: Aporrectodea rosea (Savigny, 1826), Aporrectodea caliginosa (Savigny, 1826), Aporrectodea trapezoides, Lumbricus rubellus Hoffmeister, 1843, Lumbricus terrestris Linnaeus, 1758 and Octolasion lacteum (Örley, 1881). The most common peregrine earthworm taxa of Turkey are: Aporrectodea caliginosa (Savigny, 1826), Aporrectodea rosea (Savigny, 1826), Dendrobaena veneta veneta (Rosa, 1884), Eiseniella tetraedra (Savigny, 1826), and Lumbricus rubellus Hoffmeister, 1843. INTRODUCTION The earthworm fauna of Bulgaria remains unexplored. Rosa (1897) was the first who published data on the Bulgarian earthworms. His work was followed by Černosvitov (1934, 1937), Plisko (1963), Mihailova (1964, 1965, 1966, 1968), Zicsi and Csuzdi (1986), Duhlinska (1988), Kvavadze and Miloikova (1991), Šapkarev (1986), Deltchev et al. (1998). Recently Stojanović et al. (2012, 2013), 45 Valchovski (2012, 2014), Szederjesi (2013), Valchovski and Szederjesi (2016) elaborate knowledge of earthworm fauna of the country. -

An Integrative Taxonomic Approach to the Identification of Three New New Zealand Endemic Earthworm Species (Acanthodrilidae, Octochaetidae: Oligochaeta)
Zootaxa 2994: 21–32 (2011) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2011 · Magnolia Press ISSN 1175-5334 (online edition) An integrative taxonomic approach to the identification of three new New Zealand endemic earthworm species (Acanthodrilidae, Octochaetidae: Oligochaeta) STEPHANE BOYER1,3, ROBERT J. BLAKEMORE2 & STEVE D. WRATTEN1 1Bio-Protection Research Centre, Lincoln University, New Zealand 2National Museum of Science and Nature in Tokyo, Japan 3Corresponding author. E-mail: [email protected] Abstract This work adds three new species to the ca. 200 currently known from New Zealand. In Acanthodrilidae is Maoridrilus felix and in Octochaetidae are Deinodrilus gorgon and Octochaetus kenleei. All three are endemics that often have restrict- ed ranges; however, little is yet known of their distribution, ecology nor conservation status. DNA barcoding was conduct- ed, which is the first time that New Zealand endemic holotypes have been so characterized. The barcoding region COI (cytochrome c oxidase subunit 1) as well as the 16S rDNA region were sequenced using tissue from the holotype specimen to provide indisputable uniqueness of the species. These DNA sequences are publically available on GenBank to allow accurate cross checking to verify the identification of other specimens or even to identify specimens on the basis of their DNA sequences alone. Based on their 16S rDNA sequences, the position of the three newly described species in the phy- logeny of New Zealand earthworms was discussed. The description of new species using this approach is encouraged, to provide a user-friendly identification tool for ecologists studying diverse endemic faunas of poorly known earthworm species.