Spike-Specific Circulating T Follicular Helper Cell and Cross-Neutralizing Antibody Responses in COVID-19-Convalescent Individuals
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

COVID-19 Natural Immunity
COVID-19 natural immunity Scientific brief 10 May 2021 Key Messages: • Within 4 weeks following infection, 90-99% of individuals infected with the SARS-CoV-2 virus develop detectable neutralizing antibodies. • The strength and duration of the immune responses to SARS-CoV-2 are not completely understood and currently available data suggests that it varies by age and the severity of symptoms. Available scientific data suggests that in most people immune responses remain robust and protective against reinfection for at least 6-8 months after infection (the longest follow up with strong scientific evidence is currently approximately 8 months). • Some variant SARS-CoV-2 viruses with key changes in the spike protein have a reduced susceptibility to neutralization by antibodies in the blood. While neutralizing antibodies mainly target the spike protein, cellular immunity elicited by natural infection also target other viral proteins, which tend to be more conserved across variants than the spike protein. The ability of emerging virus variants (variants of interest and variants of concern) to evade immune responses is under investigation by researchers around the world. • There are many available serologic assays that measure the antibody response to SARS-CoV-2 infection, but at the present time, the correlates of protection are not well understood. Objective of the scientific brief This scientific brief replaces the WHO Scientific Brief entitled “’Immunity passports’ in the context of COVID-19”, published 24 April 2020.1 This update is focused on what is currently understood about SARS-CoV-2 immunity from natural infection. More information about considerations on vaccine certificates or “passports”will be covered in an update of WHO interim guidance, as requested by the COVID-19 emergency committee.2 Methods A rapid review on the subject was undertaken and scientific journals were regularly screened for articles on COVID-19 immunity to ensure to include all large and robust studies available in the literature at the time of writing. -

The Urban Flood Control Project in the Mountainous Area in Hunan Province Loaned by the Asian Development Bank
The Urban Flood Control Project in the Mountainous Area in Hunan Province Loaned by the Asian Development Bank The External Resettlement Monitoring & Assessment Report (Lengshuijiang City, Lianyuan City, Shuangfeng County, Shaoyang City, Shaodong County, Longhui County, Jiangyong County, Xintian County, Jianghua County, Qiyang County, Ningyuan County, Chenzhou City, Zhuzhou City, Liling City, Zhuzhou County and Youxian County) No.1, 2008 Total No. 1 Hunan Water & Electricity Consulting Corporation (HWECC) September, 2008 Approved by: Wang Hengyang Reviewed by: Long Xiachu Prepared by: Long Xiachu, Wei Riwen 2 Contents 1. Introduction 2. Project Outline 2.1 Project Outline 2.2 Resettlement Outline 3. Establishment and Operation of Resettlement Organizations 3.1 Organization Arrangement 3.2 Organization Operation 4. Project Implementation Progress 4.1 Jiangyong County 4.2 Chenzhou City 5. Resettlement Implementation Progress 5.1 Resettlement Implementation Schedule 5.2 Resettlement Policy and Compensation Standards 5.3 Progress of Land Acquisition 5.4 Progress of Resettlement Arrangement 5.5 Removal Progress of Enterprises and Institutions 5.6 Progress of Resettlement Area Construction 5.7 Arrival and Payment of the Resettlement Fund 6. Psychology and Complaint of the Resettled People 6.1 Complaint Channel 6.2 Complaint Procedures 7. Public Participation, Consultation and Information Publicizing 7.1 Jiangyong County 7.2 Chenzhou City 8. Existed Problems and Suggestions 3 1. Introduction The Urban Flood Control Project in the Mountainous -

Anti-SARS-Cov-2 Neutralizing Antibodies
August 28, 2020 Edition 2020-08-28 (43) *** Available on-line at https://www.cdc.gov/library/covid19 *** Anti-SARS-CoV-2 Neutralizing Antibodies Anti-SARS-CoV-2 neutralizing antibodies (NAbs) can be found in persons who have recovered from COVID-19. Characterizing NAb activity might provide relevant data for understanding NAbs levels needed for natural protection against reinfection. It could also help determine the optimal design and dosing of vaccines. PEER-REVIEWED A. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in Shanghai, China. Wu et al. JAMA Internal Medicine (August 18, 2020). Key findings: • SARS-CoV-2-specific neutralizing antibody (Nab) titers varied substantially, including less than the detectable level of the assay (Figure 1). • NAbs were detected from day 4 to day 6 after symptom onset and peaked at day 10 to day 15. • NAb titers were significantly correlated with levels of spike-binding antibody and plasma C-reactive protein. • NAb titers were significantly higher in men compared with women (p = 0.01) and in middle-aged (40-59 years) and older adults (60-85 years) compared with persons 15-39 years, p <0.001 (Figure 2). Methods: Cohort study of 175 patients with laboratory-confirmed mild COVID-19 hospitalized from January 24 to February 26, 2020 at a single hospital in Shanghai, China. Plasma was tested for SARS-CoV-2–specific NAbs titers and virus spike-binding antibodies every 2 to 4 days from admission until discharge and then two weeks after discharge. Limitations: Single setting; results might not be generalizable. -

Coevolution of HIV-1 and Broadly Neutralizing Antibodies
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Curr Opin Manuscript Author HIV AIDS. Author Manuscript Author manuscript; available in PMC 2020 October 13. Published in final edited form as: Curr Opin HIV AIDS. 2019 July ; 14(4): 286–293. doi:10.1097/COH.0000000000000550. Coevolution of HIV-1 and broadly neutralizing antibodies Nicole A. Doria-Rosea, Elise Landaisb aVaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland bIAVI Neutralizing Antibody Center, Immunology and Microbiology Department, The Scripps Research Institute, La Jolla, California, USA Abstract Purpose of review—Exploring the molecular details of the coevolution of HIV-1 Envelope with broadly neutralizing antibodies (bNAbs) in infected individuals over time provides insights for vaccine design. Since mid-2017, the number of individuals described in such publications has nearly tripled. New publications have extended such studies to new epitopes on Env and provided more detail on previously known sites. Recent findings—Studies of two donors – one of them an infant, the other with three lineages targeting the same site – has deepened our understanding of V3-glycan-directed lineages. A V2- apex-directed lineage showed remarkable similarity to a lineage from a previously described donor, revealing general principles for this class of bNAbs. Understanding development of CD4 binding site antibodies has been enriched by the study of a VRC01-class lineage. Finally, the membrane-proximal external region is a new addition to the set of epitopes studied in this manner, with early development events explored in a study of three lineages from a single donor. -

Rapid Induction of Antigen-Specific CD4+ T Cells Guides Coordinated Humoral and Cellular Immune Responses to SARS-Cov-2 Mrna Vaccination
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.21.440862; this version posted April 22, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Rapid induction of antigen-specific CD4+ T cells guides coordinated humoral and cellular immune responses to SARS-CoV-2 mRNA vaccination Authors: Mark M. Painter1,2, †, Divij Mathew1,2, †, Rishi R. Goel1,2, †, Sokratis A. Apostolidis1,2,3, †, Ajinkya Pattekar2, Oliva Kuthuru1, Amy E. Baxter1, Ramin S. Herati4, Derek A. Oldridge1,5, Sigrid Gouma6, Philip Hicks6, Sarah Dysinger6, Kendall A. Lundgreen6, Leticia Kuri-Cervantes1,6, Sharon Adamski2, Amanda Hicks2, Scott Korte2, Josephine R. Giles1,7,8, Madison E. Weirick6, Christopher M. McAllister6, Jeanette Dougherty1, Sherea Long1, Kurt D’Andrea1, Jacob T. Hamilton2,6, Michael R. Betts1,6, Paul Bates6, Scott E. Hensley6, Alba Grifoni9, Daniela Weiskopf9, Alessandro Sette9, Allison R. Greenplate1,2, E. John Wherry1,2,7,8,* Affiliations 1 Institute for Immunology, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA 2 Immune Health™, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA 3 Division of Rheumatology, University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA 4 NYU Langone Vaccine Center, Department of Medicine, New York University School of Medicine, New York, NY 5 Department -

Cui Chenzhou, Zhao Yongheng, CAO Zihuang, Kubánek Petr
Curriculum Vitae Chenzhou CUI National Astronomical Observatories, Chinese Academy of Sciences (NAOC) 20A Datun Road, Chaoyang District Beijing 100012, China Tel: 86-10-64872500 Fax: 86-10-64878240 Email: [email protected] http://www.lamost.org/~cb/ Personal Information: Gender: Male Date of Birth: January 27, 1976 (Chinese Lunar Calendar) Family Status: Married, one child Nationality: P. R. of China Current Position: Chief Information Officer, Professor, NAOC, 2006.1- Education: Ph.D. in Astrophysics, NAOC, Beijing, 2003.6 Concentrations: Virtual Observatory, Grid Technology, Web Services, Scientific Databases Thesis: System Design of Chinese Virtual Observatory Advisor: Prof. Yongheng ZHAO M.Sc. in Astrophysics, Beijing Astronomical Observatory, Beijing, 2000.6 Concentrations: Astronmical Databasese, Galactic Abundances Thesis: Setup of SAGE Astronomical Database and Statistical Analyses of Galactic Abundances Advisor: Prof. Gang ZHAO B.S.E. in Mechanical Engineering, Hebei University of Technology, Tianjin, 1997.6 Research Interests: Astro-Informatics, Virtual Observatory, Grid Technology, Astronomical Databases, Astronomical Data Center, Data Processing and Analysis, Galactic Structure and Evolution, Stellar Abundance Work Experiences: Professor, NAOC, 2013.2 - Associated Professor, NAOC, 2006.1- 2013.1 Lecturer, NAOC, 2003.6-2005.12 Visiting Experiences: Visiting Scientist, Johns Hopkins University, USA, 2007.11.26-2007.12.24 Lecture, International School for Young Astronomers 2007, International Astronomical Union (IAU), Malaysia, -

SARS-Cov-2 Igg/Neutralizing Antibody Rapid Test Kit (Colloidal Gold) Instructions for Use (IFU)
accuracy. SARS-CoV-2 IgG/Neutralizing antibody Rapid Test Kit 7. Performing the assay outside the prescribed time and temperature ranges may produce invalid results. Assays not falling within the established time and temperature ranges must be repeated. (Colloidal Gold) 8. The components in this kit have been quality control tested as a master lot unit. Do not mix components from different lot numbers. Do not mix with components from other manufacturers. Instructions for Use (IFU) 9. Care should be exercised to protect the reagents in this kit from contamination. Do not use if there is evidence of microbial 【PRODUCT NAME】 contamination or precipitation. Biological contamination of dispensing equipment, containers or reagents can lead to false results. Do SARS-CoV-2 IgG/Neutralizing antibody Rapid Test Kit (Colloidal Gold) not heat-inactivate samples. 【PACKAGE AND SPECIFICATION】 10. Keep storage boxes dry. 11. Do not use test cassettes if foil pouch is punctured or damaged. 20Tests/box (1Test ×20) 、40 Tests /box (1Test ×40) 12. Testing materials should be disposed of in accordance with local, state and/or federal regulations. 【INTENDED USE】 13. Do not use after expiration date. For in vitro qualitative detect of human IgG antibodies against SARS-CoV-2 and neutralizing antibodies that block the interaction between the 14. Please read the instructions carefully before operation and follow the instructions. receptor binding domain of the viral spike glycoprotein (RBD) with the ACE2 cell surface receptor in serum, plasma and whole blood. This test 15. Please use fresh samples as much as possible, and avoid using samples contaminated with bacteria, hemolysis, jaundice, or excessive is only provided for use by clinical laboratories or to healthcare workers for point-of-care testing. -

Virus Evolution and Neutralization Sensitivity in an HIV-1 Subtype B' Infected Plasma Donor with Broadly Neutralizing Activi
Article Virus Evolution and Neutralization Sensitivity in an HIV-1 Subtype B0 Infected Plasma Donor with Broadly Neutralizing Activity Yuanyuan Hu †, Sen Zou † , Zheng Wang, Ying Liu, Li Ren, Yanling Hao, Shasha Sun, Xintao Hu ‡ , Yuhua Ruan, Liying Ma, Yiming Shao and Kunxue Hong * State Key Laboratory of Infectious Disease Prevention and Control, National Center for AIDS/STD Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 102206, China; [email protected] (Y.H.); [email protected] (S.Z.); [email protected] (Z.W.); [email protected] (Y.L.); [email protected] (L.R.); [email protected] (Y.H.); [email protected] (S.S.); [email protected] (X.H.); [email protected] (Y.R.); [email protected] (L.M.); [email protected] (Y.S.) * Correspondence: [email protected] † These authors contributed equally. ‡ Present address: Human Retrovirus Pathogenesis Section, Vaccine Branch, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA. Abstract: We sought to analyze the evolutionary characteristics and neutralization sensitivity of viruses in a human immunodeficiency virus type 1 (HIV-1) subtype B0 infected plasma donor with broadly neutralizing activity, which may provide information for new broadly neutralizing antibodies (bNAbs) isolation and immunogen design. A total of 83 full-length envelope genes were obtained by Citation: Hu, Y.; Zou, S.; Wang, Z.; single-genome amplification (SGA) from the patient’s plasma at three consecutive time points (2005, Liu, Y.; Ren, L.; Hao, Y.; Sun, S.; Hu, 2006, and 2008) spanning four years. In addition, 28 Env-pseudotyped viruses were constructed X.; Ruan, Y.; Ma, L.; et al. -

Neutralizing Antibody Response of Vaccinees to SARS-Cov-2 Variants
Brief Report Neutralizing Antibody Response of Vaccinees to SARS-CoV-2 Variants Gabriele Anichini 1 , Chiara Terrosi 1, Gianni Gori Savellini 1 , Claudia Gandolfo 1 , Federico Franchi 2 and Maria Grazia Cusi 1,* 1 Virology Unit, Department of Medical Biotechnologies, Siena University Hospital, 53100 Siena, Italy; [email protected] (G.A.); [email protected] (C.T.); [email protected] (G.G.S.); [email protected] (C.G.) 2 Emergency, Anesthesia and Intensive Care Unit, Department of Medicine, Surgery and Neurosciences, Siena University Hospital, 53100 Siena, Italy; [email protected] * Correspondence: [email protected] Abstract: Due to their increased transmissibility, three variants of high concern have emerged in the United Kingdom (also known as B.1.1.7 lineage or VOC-202012/01), South Africa (B.1.351 lineage), and Brazil (P1 lineage) with multiple substitutions in the spike protein. Since neutralizing antibodies elicited by vaccination are likely considered as correlates of protection for SARS-CoV-2 infection, it is important to analyze whether vaccinees with mRNA BNT162b2 are equally protected against these emerging SARS-CoV-2 variants. To this aim, we enrolled healthy subjects one month after complete vaccination with Comirnaty and evaluated the neutralizing response against the native Wuhan strain and the emerging B.1.1.7, B.1.351 and P1 lineages, by using the microneutralization assay, currently considered the gold standard test for the evaluation and detection of functional neutralizing antibodies. The most remarkable finding of this study was the significantly lower neutralizing antibody titer against B.1.351 lineage, compared to the wild-type virus. -

COVID-19: Mechanisms of Vaccination and Immunity
Review COVID-19: Mechanisms of Vaccination and Immunity Daniel E. Speiser 1,* and Martin F. Bachmann 2,3,4,* 1 Department of Oncology, University Hospital and University of Lausanne, 1066 Lausanne, Switzerland 2 International Immunology Centre, Anhui Agricultural University, Hefei 230036, China 3 Department of Rheumatology, Immunology and Allergology, Inselspital, University of Bern, 3010 Bern, Switzerland 4 Department of BioMedical Research, University of Bern, 3008 Bern, Switzerland * Correspondence: [email protected] (D.E.S.); [email protected] (M.F.B.) Received: 2 July 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Vaccines are needed to protect from SARS-CoV-2, the virus causing COVID-19. Vaccines that induce large quantities of high affinity virus-neutralizing antibodies may optimally prevent infection and avoid unfavorable effects. Vaccination trials require precise clinical management, complemented with detailed evaluation of safety and immune responses. Here, we review the pros and cons of available vaccine platforms and options to accelerate vaccine development towards the safe immunization of the world’s population against SARS-CoV-2. Favorable vaccines, used in well-designed vaccination strategies, may be critical for limiting harm and promoting trust and a long-term return to normal public life and economy. Keywords: SARS-CoV-2; COVID-19; nucleic acid tests; serology; vaccination; immunity 1. Introduction The COVID-19 pandemic holds great challenges for which the world is only partially prepared [1]. SARS-CoV-2 combines serious pathogenicity with high infectivity. The latter is enhanced by the fact that asymptomatic and pre-symptomatic individuals can transmit the virus, in contrast to SARS-CoV-1 and MERS-CoV, which were transmitted by symptomatic patients and could be contained more efficiently [2,3]. -
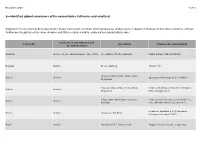
2—Identified Global Consumers of Tin Concentrates (Refineries and Smelters)
November 2020 1 of 12 2—Identified global consumers of tin concentrates (refineries and smelters) Explanation: This list contains global facilities known to be able to process tin concentrate. Additional small-scale smelters may be in operation in Indonesia, but their status could not be confirmed. Facilities were thought to be active unless otherwise noted. Data records are sorted by country and then alphabetically by owner. FACILITY TYPE AND STATUS COUNTRY LOCATION OPERATOR / OWNERSHIP (IF APPLICABLE) Australia Smelter (on care and maintenance since 2012) Greenbushes, Western Australia Global Advanced Metals Pty Ltd. Belgium Smelter Beerse, Antwerp Aurubis AG Huajara Industrial Park, Oruro, Oruro Bolivia Smelter Operaciones Metalúrgicas S.A. (OMSA) Department Vinto smelting complex, Vinto, Oruro Empresa Metalúrgica Vinto S.A. (Compania Bolivia Smelter Department Minera Colquira S.A.) Coopersanta, Bom Futuro, Ariquemes, Cooperativa de Garimperiros de Santa Cruz Brazil Smelter Rondônia Ltda. [Meridian Mineração Jaburi S.A.] Estanho de Rondônia S.A. [Companhia Brazil Smelter Ariquemes, Rondônia Siderúrgica Nacional (CSN)] Brazil Smelter São João del Rei, Minas Gerais Magnu's Minerais Metais e Ligas Ltda. November 2020 2 of 12 2—Identified global consumers of tin concentrates (refineries and smelters) Explanation: This list contains global facilities known to be able to process tin concentrate. Additional small-scale smelters may be in operation in Indonesia, but their status could not be confirmed. Facilities were thought to be active unless otherwise noted. Data records are sorted by country and then alphabetically by owner. FACILITY TYPE AND STATUS COUNTRY LOCATION OPERATOR / OWNERSHIP (IF APPLICABLE) Brazil Smelter Ariquemes, Rondônia Melt Metais e Ligas S.A. -

Escape from Neutralizing Antibodies by SARS-Cov-2 Spike Protein Variants
RESEARCH ARTICLE Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants Yiska Weisblum1†, Fabian Schmidt1†, Fengwen Zhang1, Justin DaSilva1, Daniel Poston1, Julio CC Lorenzi2, Frauke Muecksch1, Magdalena Rutkowska1, Hans-Heinrich Hoffmann3, Eleftherios Michailidis3, Christian Gaebler2, Marianna Agudelo2, Alice Cho2, Zijun Wang2, Anna Gazumyan2, Melissa Cipolla2, Larry Luchsinger4, Christopher D Hillyer4, Marina Caskey2, Davide F Robbiani2,5, Charles M Rice3, Michel C Nussenzweig2,6, Theodora Hatziioannou1*, Paul D Bieniasz1,6* 1Laboratory of Retrovirology, The Rockefeller University, New York, United States; 2Laboratory of Molecular Immunology The Rockefeller University, New York, United States; 3Laboratory of Virology and Infectious Disease The Rockefeller University, New York, United States; 4Lindsley F. Kimball Research Institute, New York Blood Center, New York, United States; 5Institute for Research in Biomedicine, Universita` della Svizzera italiana, Bellinzona, Switzerland; 6Howard Hughes Medical Institute, The Rockefeller University, New York, United States Abstract Neutralizing antibodies elicited by prior infection or vaccination are likely to be key for future protection of individuals and populations against SARS-CoV-2. Moreover, passively administered antibodies are among the most promising therapeutic and prophylactic anti-SARS- CoV-2 agents. However, the degree to which SARS-CoV-2 will adapt to evade neutralizing antibodies is unclear. Using a recombinant chimeric VSV/SARS-CoV-2 reporter virus, we show that functional SARS-CoV-2 S protein variants with mutations in the receptor-binding domain (RBD) and *For correspondence: N-terminal domain that confer resistance to monoclonal antibodies or convalescent plasma can be [email protected] (TH); [email protected] (PDB) readily selected. Notably, SARS-CoV-2 S variants that resist commonly elicited neutralizing antibodies are now present at low frequencies in circulating SARS-CoV-2 populations.