Double-Strand Signal Sequence Breaks in V(D)J Recombination Are Blunt, 5'-Phosphorylated, RAG-Dependent, and Cell Cycle Regulated
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

College Summit, Inc. D/B/A Peerforward Financial Statements
College Summit, Inc. d/b/a PeerForward Financial Statements For the Years Ended April 30, 2020 and 2019 and Report Thereon COLLEGE SUMMIT, INC. d/b/a PeerForward TABLE OF CONTENTS For the Years Ended April 30, 2020 and 2019 _______________ Page Independent Auditors’ Report ............................................................................................................. 1-2 Financial Statements Statements of Financial Position ........................................................................................................ 3 Statements of Activities ...................................................................................................................... 4 Statements of Functional Expenses ................................................................................................ 5-6 Statements of Cash Flows .................................................................................................................. 7 Notes to Financial Statements ....................................................................................................... 8-17 INDEPENDENT AUDITORS’ REPORT To the Board of Directors of College Summit, Inc. d/b/a PeerForward Report on the Financial Statements We have audited the accompanying financial statements of College Summit, Inc. d/b/a PeerForward (PeerForward), which comprise the statements of financial position as of April 30, 2020 and 2019, and the related statements of activities, functional expenses and cash flows for the years then ended, and the related notes -

How Can We Create Environments Where Hate Cannot Flourish?
How can we create environments where hate cannot flourish? Saturday, February 1 9:30 a.m. – 10:00 a.m. Check-In and Registration Location: Field Museum West Entrance 10:00 a.m. – 10:30 a.m. Introductions and Program Kick-Off JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1 10:30 a.m. – 11:30 a.m. Watch “The Path to Nazi Genocide” and Reflections JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1 11:30 a.m. – 11:45 a.m. Break and walk to State of Deception 11:45 a.m. – 12:30 p.m. Visit State of Deception Interpretation by Holocaust Survivor Volunteers from the Illinois Holocaust Museum Location: Upper level 12:30 p.m. – 1:00 p.m. Breakout Session: Reflections on Exhibit Tim Kaiser, USHMM Director, Education Initiatives David Klevan, USHMM Digital Learning Strategist JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1, Classrooms A and B Saturday, February 2 (continued) 1:00 p.m. – 1:45 p.m. Lunch 1:45 p.m. – 2:45 p.m. A Survivor’s Personal Story Bob Behr, USHMM Survivor Volunteer Interviewed by: Ann Weber, USHMM Program Coordinator Location: Lecture Hall 1 2:45 p.m. – 3:00 p.m. Break 3:00 p.m. – 3:45 p.m. Student Panel: Beyond Indifference Location: Lecture Hall 1 Moderator: Emma Pettit, Sustained Dialogue Campus Network Student/Alumni Panelists: Jazzy Johnson, Northwestern University Mary Giardina, The Ohio State University Nory Kaplan-Kelly, University of Chicago 3:45 p.m. – 4:30 p.m. Breakout Session: Sharing Personal Reflections Tim Kaiser, USHMM Director, Education Initiatives David Klevan, USHMM Digital Learning Strategist JoAnna Wasserman, USHMM Education Initiatives Manager Location: Lecture Hall 1, Classrooms A and B 4:30 p.m. -

Stroke Training for EMS Professionals (PDF)
STROKE TRAINING FOR EMS PROFESSIONALS 1 COURSE OBJECTIVES About Stroke Stroke Policy Recommendations Stroke Protocols and Stroke Hospital Care Stroke Assessment Tools Pre-Notification Stroke Treatment ABOUT STROKE STROKE FACTS • A stroke is a medical emergency! Stroke occurs when blood flow is either cut off or is reduced, depriving the brain of blood and oxygen • Approximately 795,000 strokes occur in the US each year • Stroke is the fifth leading cause of death in the US • Stroke is a leading cause of adult disability • On average, every 40 seconds, someone in the United States has a stroke • Over 4 million stroke survivors are in the US • The indirect and direct cost of stroke: $38.6 billion annually (2009) • Crosses all ethnic, racial and socioeconomic groups Berry, Jarett D., et al. Heart Disease and Stroke Statistics --2013 Update: A Report from the American Heart Association. Circulation. 127, 2013. DIFFERENT TYPES OF STROKE Ischemic Stroke • Caused by a blockage in an artery stopping normal blood and oxygen flow to the brain • 87% of strokes are ischemic • There are two types of ischemic strokes: Embolism: Blood clot or plaque fragment from elsewhere in the body gets lodged in the brain Thrombosis: Blood clot formed in an artery that provides blood to the brain Berry, Jarett D., et al. Heart Disease and Stroke Statistics --2013 Update: A Report from the American Heart Association. Circulation. 127, 2013. http://www.strokeassociation.org/STROKEORG/AboutStroke/TypesofStroke/IschemicClots/Ischemic-Strokes- Clots_UCM_310939_Article.jsp -

Medical Terminology Abbreviations Medical Terminology Abbreviations
34 MEDICAL TERMINOLOGY ABBREVIATIONS MEDICAL TERMINOLOGY ABBREVIATIONS The following list contains some of the most common abbreviations found in medical records. Please note that in medical terminology, the capitalization of letters bears significance as to the meaning of certain terms, and is often used to distinguish terms with similar acronyms. @—at A & P—anatomy and physiology ab—abortion abd—abdominal ABG—arterial blood gas a.c.—before meals ac & cl—acetest and clinitest ACLS—advanced cardiac life support AD—right ear ADL—activities of daily living ad lib—as desired adm—admission afeb—afebrile, no fever AFB—acid-fast bacillus AKA—above the knee alb—albumin alt dieb—alternate days (every other day) am—morning AMA—against medical advice amal—amalgam amb—ambulate, walk AMI—acute myocardial infarction amt—amount ANS—automatic nervous system ant—anterior AOx3—alert and oriented to person, time, and place Ap—apical AP—apical pulse approx—approximately aq—aqueous ARDS—acute respiratory distress syndrome AS—left ear ASA—aspirin asap (ASAP)—as soon as possible as tol—as tolerated ATD—admission, transfer, discharge AU—both ears Ax—axillary BE—barium enema bid—twice a day bil, bilateral—both sides BK—below knee BKA—below the knee amputation bl—blood bl wk—blood work BLS—basic life support BM—bowel movement BOW—bag of waters B/P—blood pressure bpm—beats per minute BR—bed rest MEDICAL TERMINOLOGY ABBREVIATIONS 35 BRP—bathroom privileges BS—breath sounds BSI—body substance isolation BSO—bilateral salpingo-oophorectomy BUN—blood, urea, nitrogen -
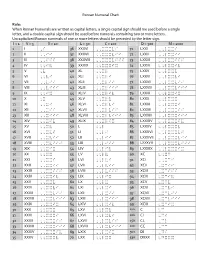
Roman Numeral Chart
Roman Numeral Chart Rule: When Roman Numerals are written as capital letters, a single capital sign should me used before a single letter, and a double capital sign should be used before numerals containing two or more letters. Uncapitalized Roman numerals of one or more letters should be preceded by the letter sign. I = 1 V = 5 X = 10 L = 50 C = 100 D = 500 M = 1000 1 I ,I 36 XXXVI ,,XXXVI 71 LXXI ,,LXXI 2 II ,,II 37 XXXVII ,,XXXVII 72 LXXII ,,LXXII 3 III ,,III 38 XXXVIII ,,XXXVIII 73 LXXIII ,,LXXIII 4 IV ,,IV 39 XXXIX ,,XXXIX 74 LXXIV ,,LXXIV 5 V ,V 40 XL ,,XL 75 LXXV ,,LXXV 6 VI ,,VI 41 XLI ,,XLI 76 LXXVI ,,LXXVI 7 VII ,,VII 42 XLII ,,XLII 77 LXXVII ,,LXXVII 8 VIII ,,VIII 43 XLIII ,,XLIII 78 LXXVIII ,,LXXVIII 9 IX ,,IX 44 XLIV ,,XLIV 79 LXXIX ,,LXXIX 10 X ,X 45 XLV ,,XLV 80 LXXX ,,LXXX 11 XI ,,XI 46 XLVI ,,XLVI 81 LXXXI ,,LXXXI 12 XII ,,XII 47 XLVII ,,XLVII 82 LXXXII ,,LXXXII 13 XIII ,,XIII 48 XLVIII ,,XLVIII 83 LXXXIII ,,LXXXIII 14 XIV ,,XIV 49 XLIX ,,XLIX 84 LXXXIV ,,LXXXIV 15 XV ,,XV 50 L ,,L 85 LXXXV ,,LXXXV 16 XVI ,,XVI 51 LI ,,LI 86 LXXXVI ,,LXXXVI 17 XVII ,,XVII 52 LII ,,LII 87 LXXXVII ,,LXXXVII 18 XVIII ,,XVIII 53 LIII ,,LIII 88 LXXXVIII ,,LXXXVIII 19 XIX ,,XIX 54 LIV ,,LIV 89 LXXXIX ,,LXXXIX 20 XX ,,XX 55 LV ,,LV 90 XC ,,XC 21 XXI ,,XXI 56 LVI ,,LVI 91 XCI ,,XCI 22 XXII ,,XXII 57 LVII ,,LVII 92 XCII ,XCII 23 XXIII ,,XXIII 58 LVIII ,,LVIII 93 XCIII ,XCIII 24 XXIV ,,XXIV 59 LIX ,,LIX 94 XCIV ,XCIV 25 XXV ,,XXV 60 LX ,,LX 95 XCV ,XCV 26 XXVI ,,XXVI 61 LXI ,,LXI 96 XCVI ,XCVI 27 XXVII ,,XXVII 62 LXII ,,LXII 97 XCVII ,XCVII 28 XXVIII ,,XXVIII 63 LXIII ,,LXIII 98 XCVIII ,XCVIII 29 XXIX ,,XXIX 64 LXIV ,,LXIV 99 XCIX ,XCIX 30 XXX ,,XXX 65 LXV ,,LXV 100 C ,C 31 XXXI ,,XXXI 66 LXVI ,,LXVI 101 CI ,CI 32 XXXII ,,XXXII 67 LXVII ,,LXVII 150 CL ,CL 33 XXXIII ,,XXXIII 68 LXVIII ,,LXVIII 200 CC ,CC 34 XXXIV ,,XXXIV 69 LXIX ,,LXIX 501 DI ,DI 35 XXXV ,,XXXV 70 LXX ,,LXX 530 DXXX ,DXXX . -

List of Approved Special Characters
List of Approved Special Characters The following list represents the Graduate Division's approved character list for display of dissertation titles in the Hooding Booklet. Please note these characters will not display when your dissertation is published on ProQuest's site. To insert a special character, simply hold the ALT key on your keyboard and enter in the corresponding code. This is only for entering in a special character for your title or your name. The abstract section has different requirements. See abstract for more details. Special Character Alt+ Description 0032 Space ! 0033 Exclamation mark '" 0034 Double quotes (or speech marks) # 0035 Number $ 0036 Dollar % 0037 Procenttecken & 0038 Ampersand '' 0039 Single quote ( 0040 Open parenthesis (or open bracket) ) 0041 Close parenthesis (or close bracket) * 0042 Asterisk + 0043 Plus , 0044 Comma ‐ 0045 Hyphen . 0046 Period, dot or full stop / 0047 Slash or divide 0 0048 Zero 1 0049 One 2 0050 Two 3 0051 Three 4 0052 Four 5 0053 Five 6 0054 Six 7 0055 Seven 8 0056 Eight 9 0057 Nine : 0058 Colon ; 0059 Semicolon < 0060 Less than (or open angled bracket) = 0061 Equals > 0062 Greater than (or close angled bracket) ? 0063 Question mark @ 0064 At symbol A 0065 Uppercase A B 0066 Uppercase B C 0067 Uppercase C D 0068 Uppercase D E 0069 Uppercase E List of Approved Special Characters F 0070 Uppercase F G 0071 Uppercase G H 0072 Uppercase H I 0073 Uppercase I J 0074 Uppercase J K 0075 Uppercase K L 0076 Uppercase L M 0077 Uppercase M N 0078 Uppercase N O 0079 Uppercase O P 0080 Uppercase -

Proposal for Generation Panel for Latin Script Label Generation Ruleset for the Root Zone
Generation Panel for Latin Script Label Generation Ruleset for the Root Zone Proposal for Generation Panel for Latin Script Label Generation Ruleset for the Root Zone Table of Contents 1. General Information 2 1.1 Use of Latin Script characters in domain names 3 1.2 Target Script for the Proposed Generation Panel 4 1.2.1 Diacritics 5 1.3 Countries with significant user communities using Latin script 6 2. Proposed Initial Composition of the Panel and Relationship with Past Work or Working Groups 7 3. Work Plan 13 3.1 Suggested Timeline with Significant Milestones 13 3.2 Sources for funding travel and logistics 16 3.3 Need for ICANN provided advisors 17 4. References 17 1 Generation Panel for Latin Script Label Generation Ruleset for the Root Zone 1. General Information The Latin script1 or Roman script is a major writing system of the world today, and the most widely used in terms of number of languages and number of speakers, with circa 70% of the world’s readers and writers making use of this script2 (Wikipedia). Historically, it is derived from the Greek alphabet, as is the Cyrillic script. The Greek alphabet is in turn derived from the Phoenician alphabet which dates to the mid-11th century BC and is itself based on older scripts. This explains why Latin, Cyrillic and Greek share some letters, which may become relevant to the ruleset in the form of cross-script variants. The Latin alphabet itself originated in Italy in the 7th Century BC. The original alphabet contained 21 upper case only letters: A, B, C, D, E, F, Z, H, I, K, L, M, N, O, P, Q, R, S, T, V and X. -

Michael D. O'mara
FOCUS Michael D. O’Mara Litigation Complex Commercial & Class Chair, Litigation Action Litigation Philadelphia, PA Real Estate Litigation 215.564.8121 BAR ADMISSIONS [email protected] Pennsylvania New Jersey New York COURTS U.S. Court of Appeals for the Fourth Circuit Mike O’Mara is a recognized trial lawyer who focuses his practice on complex and U.S. Court of Appeals for the contentious commercial disputes. He chairs Stradley Ronon’s 65-attorney Third Circuit litigation department and also serves on the firm’s board of directors. U.S. District Court for the Eastern District of Pennsylvania Mike’s experience covers a broad spectrum of commercial litigation, including U.S. District Court for the Middle District financial services cases, merger and acquisition disputes, intellectual property of Pennsylvania litigation, class actions, ERISA claims, trust and estate disputes, and insurance and U.S. District Court for the District of commercial real estate matters. He frequently represents professional services New Jersey firms – including Am Law 100 and 200 firms – in professional liability cases. Mike also counsels a number of nonprofit organizations, including religious, EDUCATION educational and health care organizations, on a wide variety of litigation-related J.D., cum laude, Pennsylvania State matters. University Dickinson School of Law He routinely tries cases in federal and state courts, as well as before private B.S., magna cum laude , James Madison University arbitrators. He was selected to join the Litigation Counsel of America, a prestigious invitation-only trial lawyer honorary society open to just one-half of MEMBERSHIPS one percent of U.S. lawyers. He also serves as an adjunct faculty member at Villanova School of Law, where he teaches a class on trial advocacy. -

MCI Communications Services LLC D/B/A Verizon Business Services Arizona Tariff No
MCI Communications Services LLC d/b/a Verizon Business Services Arizona Tariff No. 1 (T) Price List 1st Revised Sheet No. PL - 1 Cancels Original Sheet No. PL - 1 INTRASTATE TELECOMMUNICATIONS SERVICES PRICE LIST 1. WorldOne Service Per Minute Rates 1.1 Switched Per Minute Maximum Rates Monthly 1 Year ESP 2 Year ESP Outbound $.1500 $.1450 $.1400 Inbound $.1600 $.1450 $.1400 1.2 Dedicated Per Minute Maximum Rates Monthly 1 Year ESP 2 Year ESP Outbound $.1100 $.1050 $.1000 Inbound $.1200 $.1050 $.1000 1.3 OnLine World Calling Card Service Per minute rate $0.3000 Per call surcharge $0.65 1.3.1 Calls which default to a live operator: Rate Per Minute: Peak $0.23 Off-Peak $0.16 Per call Surcharge $0.65 1.3.2 OnLine World Calling Card Service - Online Operator Assisted Rate Schedule: Per Call Surcharge Station-to-Station $1.50 Person-to-Person $3.00 1.4 WorldOne Association Monthly ESP Switched Outbound $0.1450 $0.1400 Switched Toll Free $0.1450 $0.1400 Dedicated Outbound $0.1050 $0.1000 Dedicated Toll Free $0.1050 $0.1000 Issued: May 28, 2020 Effective: July 1, 2020 Edwin Reese Analyst-Govt Relations 1300 I Street NW, Suite 500E Washington, DC 20005 MCI Communications Services LLC d/b/a Verizon Business Services Arizona Tariff No. 1 (T) Price List 2nd Revised Sheet No. PL - 2 Cancels 1st Revised Sheet No. PL - 2 INTRASTATE TELECOMMUNICATIONS SERVICES PRICE LIST 2. [Reserved For Future Use.] Issued: May 28, 2020 Effective: July 1, 2020 Edwin Reese Analyst-Govt Relations 1300 I Street NW, Suite 500E Washington, DC 20005 MCI Communications Services LLC d/b/a Verizon Business Services Arizona Tariff No. -

Z Wkzd^ E &/E E / >^D D D Ed^ &Kzd, D, &/^ > Z Z E D Z KZ /E Zz^, Z
5 ; 5 5 88 8 5 5 5 5 8 7 ; 8888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 88 88888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 8888888888888888888888888888888888888888888888888888888888888 8888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 ; ; 88888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 , 88888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 , 88 8 888 ; 5 8888888888888888888888888 5 88 888888888888888 ;. 88 8888888888888888888888888888888888888888888888888888 88 8888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 . 888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 ,88888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888888 8888888888888888888888888888888888888888888 5 -

Fonts for Latin Paleography
FONTS FOR LATIN PALEOGRAPHY Capitalis elegans, capitalis rustica, uncialis, semiuncialis, antiqua cursiva romana, merovingia, insularis majuscula, insularis minuscula, visigothica, beneventana, carolina minuscula, gothica rotunda, gothica textura prescissa, gothica textura quadrata, gothica cursiva, gothica bastarda, humanistica. User's manual 5th edition 2 January 2017 Juan-José Marcos [email protected] Professor of Classics. Plasencia. (Cáceres). Spain. Designer of fonts for ancient scripts and linguistics ALPHABETUM Unicode font http://guindo.pntic.mec.es/jmag0042/alphabet.html PALEOGRAPHIC fonts http://guindo.pntic.mec.es/jmag0042/palefont.html TABLE OF CONTENTS CHAPTER Page Table of contents 2 Introduction 3 Epigraphy and Paleography 3 The Roman majuscule book-hand 4 Square Capitals ( capitalis elegans ) 5 Rustic Capitals ( capitalis rustica ) 8 Uncial script ( uncialis ) 10 Old Roman cursive ( antiqua cursiva romana ) 13 New Roman cursive ( nova cursiva romana ) 16 Half-uncial or Semi-uncial (semiuncialis ) 19 Post-Roman scripts or national hands 22 Germanic script ( scriptura germanica ) 23 Merovingian minuscule ( merovingia , luxoviensis minuscula ) 24 Visigothic minuscule ( visigothica ) 27 Lombardic and Beneventan scripts ( beneventana ) 30 Insular scripts 33 Insular Half-uncial or Insular majuscule ( insularis majuscula ) 33 Insular minuscule or pointed hand ( insularis minuscula ) 38 Caroline minuscule ( carolingia minuscula ) 45 Gothic script ( gothica prescissa , quadrata , rotunda , cursiva , bastarda ) 51 Humanist writing ( humanistica antiqua ) 77 Epilogue 80 Bibliography and resources in the internet 81 Price of the paleographic set of fonts 82 Paleographic fonts for Latin script 2 Juan-José Marcos: [email protected] INTRODUCTION The following pages will give you short descriptions and visual examples of Latin lettering which can be imitated through my package of "Paleographic fonts", closely based on historical models, and specifically designed to reproduce digitally the main Latin handwritings used from the 3 rd to the 15 th century. -

Spherical Coordinates
Spherical Coordinates z r^ Transforms ^ " r The forward and reverse coordinate transformations are ! ^ ! r = x2 + y2 + z 2 r x = rsin! cos" y arctan" x2 y2 , z$ y r sin sin ! = # + % = ! " z = rcos! & = arctan(y,x) x " where we formally take advantage of the two argument arctan function to eliminate quadrant confusion. Unit Vectors The unit vectors in the spherical coordinate system are functions of position. It is convenient to express them in terms of the spherical coordinates and the unit vectors of the rectangular coordinate system which are not themselves functions of position. ! r xxˆ + yyˆ + zzˆ rˆ = = = xˆ sin! cos" + yˆ sin! sin " + zˆ cos! r r zˆ #rˆ "ˆ = = $ xˆ sin " + yˆ cos" sin! !ˆ = "ˆ # rˆ = xˆ cos! cos" + yˆ cos! sin " $ zˆ sin! Variations of unit vectors with the coordinates Using the expressions obtained above it is easy to derive the following handy relationships: !rˆ = 0 !r !rˆ = xˆ cos" cos# + yˆ cos" sin # $ zˆ sin " = "ˆ !" !rˆ = $xˆ sin" sin # + yˆ sin " cos# = ($ xˆ sin # + yˆ cos #)sin" = #ˆ sin" !# !"ˆ = 0 !r !"ˆ = 0 !# !"ˆ = $xˆ cos" $ yˆ sin " = $ rˆ sin # + #ˆ cos# !" ( ) !"ˆ = 0 !r !"ˆ = # xˆ sin " cos$ # yˆ sin" sin $ # zˆ cos" = #rˆ !" !"ˆ = # xˆ cos" sin $ + yˆ cos" cos$ = $ˆ cos" !$ Path increment ! We will have many uses for the path increment d r expressed in spherical coordinates: ! $ !rˆ !rˆ !rˆ ' dr = d(rrˆ ) = rˆ dr + rdrˆ = rˆ dr + r& dr + d" + d#) % !r !" !# ( = rˆ dr +"ˆ rd " + #ˆr sin "d# Time derivatives of the unit vectors We will also have many uses for the time derivatives of the unit vectors