Viral Pathogens and Microbial Indicators in the Canals of the Florida
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Florida Keys Vessel Pumpout Facilities Marine Sanitation Device
Marine Sanitation Device Discharge Regulations Effective: December 27, 2010 Activities prohibited Sanctuary-Wide: q Discharge of sewage incidental to vessel use and generated by a marine sanitation device in accordance with the Federal Water Pollution Control Act (also called the Clean Water Act). q Having a marine sanitation device that is not secured in a manner that prevents discharges or deposits of treated and untreated sewage. Acceptable methods include, but are not limited to, all methods that have been approved by the U.S. Coast Guard. Pumpout facilities are located throughout the Keys to assist boat operators in complying with this rule. For a list of pumpout facilities, visit http://www.dep.state.fl.us/cleanmarina/about.htm. Florida Keys Vessel Pumpout Facilities * Designated Clean Marina Facility Key West Duck Key Mobile Pumpout Services • A & B Marina • Hawk’s Cay Resort Marina Free pumpout services for vessels • Conch Harbor Marina* anchored within unincorporated Long Key • City Marina at Garrison Bight* Monroe County (Key Largo, • Key West Bight Marina* • Fiesta Key KOA Tavernier, Cudjoe, Big Pine, Stock Stock Island Upper Matecumbe Key Island, etc.) and the Village of • Stock Island Marina Village • Bayside Marina- World Wide Sportsman* Islamorada. • Sunset Marina • Coral Bay Marina • Pumpout USA at 305-900-0263 or visit www.po-keys.com. Lower Keys Plantation Key • Plantation Yacht Harbor* • Bahia Honda State Park* • City of Key West 305-292-8167 • Sunshine Key RV Resort & Marina • Treasure Harbor Marine* • Stock Island, Mark LPS 305-587-2787 Marathon Tavernier • City of Marathon 305-289-8877 • Boot Key Harbor City Marina • Mangrove Marina • Key Colony Beach 305-289-1310 • Burdines Waterfront • Marathon Yacht Club Key Largo • Panchos Fuel Dock & Marina • All Keys Portalet Tips: • Sombrero Marina Dockside* • John Pennekamp Coral Reef State Park* • Check with marina ahead of time on • Manatee Bay Marina status of pumpout equipment. -

USGS 7.5-Minute Image Map for Plantation Key, Florida
U.S. DEPARTMENT OF THE INTERIOR PLANTATION KEY QUADRANGLE U.S. GEOLOGICAL SURVEY FLORIDA-MONROE CO. ATLANTIC 7.5-MINUTE SERIES 80°37'30" East Key 35' OCEAN 32'30" 4 80°30' 5 000m 5 5 5 5 5 5 5 5 5T63S R38E 5 5 820 000 FEET 5 25°00' 38 E 39 40 41 43 44 45 46 47 48 49 50 25°00' Crane East Key Plantation Key Colony PLANTATION DR Crane Tavernier Creek Keys Keys 10 OCEAN DR T62S R38E ¤£1 4 Tavernier35 Key 2 F HIBISCUS ST ONTAINE DR JASMINE ST F GARDENIA ST T63S R38E 5 WOODS AVE Y W n█ H Kalteux Key 240 000 D D L 5 T63S n█R38EO FEET 64000m27 N «¬5 2764 EVERGLADES ATLANTIC OJIBWAY AVE 5 IND OCEAN IA NATIONAL PARK N M O U N D D Cotton CALOOSA ST T R Key L Intracoastal Basin Plantation Key Waterway 5 n█ 5 27 63 2763 8 Y HEIGHTS DR Florida KE Plantation Key Bay 10 5 Plantation 27 62 27 7 62 D LV B N O ATI PLANT Y W H D D L O 18 27 61 27 10 61 ¤£1 16 16 Cotton Imagery................................................NAIP, January 2010 Snake V ¬5 E « Key N Roads..............................................©2006-2010 Tele Atlas E Cotton Creek T 13 n█ IA Names...............................................................GNIS, 2010 57'Key30" N 57'30" Hydrography.................National Hydrography Dataset, 2010 ATLANTIC 14 B L V 5 Contours............................National Elevation Dataset, 2010 OCEAN D E RIDGE RD Plantation 5 Treasure Point 27 24 Harbor 60 2760 10 5 23 T63S R37E 5 Windley Key 27 59 2759 5 Windley Y Wilson HW Harbor OL D Key The Rocks O 22 ROE C «¬5 MON Whale ¤£1 Harbor 21 Whale Harbor 5 Channel 27 n 58 █ 2758 Hen and Chickens Upper -

Hydrographic / Bathymetric Surveys of Keys Canals
FLORIDA CITY ISLANDIA Appendix A NORTH KEY LARGO 1 in = 7 miles +/- FLAMINGO KEY LARGO TAVERNIER PLANTATION KEY ISLAMORADA LAYTON MARATHON CUDJOE KEY BIG PINE KEY SUMMERLAND KEY BIG COPPITT KEY KEY WEST Table 1 Monroe County Residential Canal Inventory Sort_Ind Canal_Name Canals Not Surveyed in S0640 Grant 1 1 OCEAN REEF CLUB 2 2 OCEAN REEF CLUB Not surveyed, is a natural channel 3 2 OCEAN REEF CLUB ADDED 4 2 OCEAN REEF CLUB ADDED 2 Not surveyed, is a natural channel 5 3 OCEAN REEF CLUB 6 4 OCEAN REEF CLUB 7 5 OCEAN REEF CLUB 8 6 KEY LARGO 9 7 KEY LARGO 10 8 KEY LARGO 11 9 KEY LARGO 12 10 KEY LARGO 13 11 KEY LARGO 14 12 KEY LARGO 15 13 KEY LARGO 16 14 KEY LARGO 17 15 KEY LARGO 18 16 KEY LARGO 19 17 KEY LARGO 20 18 KEY LARGO 21 19 KEY LARGO Not surveyed, inaccessible due to earthen dam 22 20 KEY LARGO Not surveyed, inaccessible due to earthen dam 23 21 KEY LARGO 24 22 KEY LARGO 25 23 KEY LARGO 26 24 KEY LARGO 27 25 KEY LARGO 28 26 KEY LARGO 29 27 KEY LARGO 30 28 KEY LARGO 31 29 KEY LARGO 32 30 KEY LARGO 33 31 KEY LARGO 34 32 KEY LARGO 35 33 KEY LARGO 36 34 KEY LARGO 37 35 KEY LARGO 38 36 KEY LARGO 39 37 KEY LARGO 40 38 KEY LARGO 41 39 KEY LARGO 42 40 KEY LARGO 43 41 KEY LARGO 44 42 KEY LARGO 45 43 KEY LARGO 46 44 KEY LARGO 47 45 KEY LARGO 48 46 KEY LARGO 49 47 KEY LARGO 50 48 KEY LARGO 51 49 KEY LARGO 52 50 KEY LARGO 53 51 KEY LARGO 54 52 KEY LARGO 55 53 KEY LARGO 56 54 KEY LARGO 57 55 KEY LARGO 58 56 KEY LARGO 59 57 KEY LARGO 60 58 KEY LARGO 61 59 KEY LARGO 62 60 KEY LARGO 63 61 KEY LARGO 64 62 ROCK HARBOR Not surveyed, inaccessible, -

Mile Marker 0-65 (Lower Keys)
Key to Map: Map is not to scale Existing Florida Keys Overseas Heritage Trail Aquatic Preserves or Alternate Path Overseas Paddling Trail U.S. 1 Point of Interest U.S. Highway 1 TO MIAMI Kayak/Canoe Launch Site CARD SOUND RD Additional Paths and Lanes TO N KEY LARGO Chamber of Commerce (Future) Trailhead or Rest Area Information Center Key Largo Dagny Johnson Trailhead Mangroves Key Largo Hammock Historic Bridge-Fishing Botanical State Park Islands Historic Bridge Garden Cove MM Mile Marker Rattlesnake Key MM 105 Florida Department of Environmental Protection, Office of Greenways & Trails Florida Keys Overseas Heritage Trail Office: (305) 853-3571 Key Largo Adams Waterway FloridaGreenwaysAndTrails.com El Radabob Key John Pennekamp Coral Reef State Park MM 100 Swash Friendship Park Keys Key Largo Community Park Florida Keys Community of Key Largo FLORIDA BAY MM 95 Rodriguez Key Sunset Park Dove Key Overseas Heritage Trail Town of Tavernier Harry Harris Park Burton Drive/Bicycle Lane MM 90 Tavernier Key Plantation Key Tavernier Creek Lignumvitae Key Aquatic Preserve Founders Park ATLANTIC OCEAN Windley Key Fossil Reef Geological State Park MM 85 Snake Creek Long Key Historic Bridge TO UPPER Islamorada, Village of Islands Whale Harbor Channel GULF OF MEXICO KEYS Tom's Harbor Cut Historic Bridge Wayside Rest Area Upper Matecumbe Key Tom's Harbor Channel Historic Bridge MM 80 Dolphin Research Center Lignumvitae Key Botanical State Park Tea Table Key Relief Channel Grassy Key MM 60 Conch Keys Tea Table Channel Grassy Key Rest Area Indian Key -

BISCAYNE NATIONAL PARK the Florida Keys Begin with Soldier Key in the Northern Section of the Park and Continue to the South and West
CHAPTER TWO: BACKGROUND HISTORY GEOLOGY AND PHYSICAL GEOGRAPHY OF BISCAYNE NATIONAL PARK The Florida Keys begin with Soldier Key in the northern section of the Park and continue to the south and west. The upper Florida Keys (from Soldier to Big Pine Key) are the remains of a shallow coral patch reef that thrived one hundred thousand or more years ago, during the Pleistocene epoch. The ocean level subsided during the following glacial period, exposing the coral to die in the air and sunlight. The coral was transformed into a stone often called coral rock, but more correctly termed Key Largo limestone. The other limestones of the Florida peninsula are related to the Key Largo; all are basically soft limestones, but with different bases. The nearby Miami oolitic limestone, for example, was formed by the precipitation of calcium carbonate from seawater into tiny oval particles (oolites),2 while farther north along the Florida east coast the coquina of the Anastasia formation was formed around the shells of Pleistocene sea creatures. When the first aboriginal peoples arrived in South Florida approximately 10,000 years ago, Biscayne Bay was a freshwater marsh or lake that extended from the rocky hills of the present- day keys to the ridge that forms the current Florida coast. The retreat of the glaciers brought about a gradual rise in global sea levels and resulted in the inundation of the basin by seawater some 4,000 years ago. Two thousand years later, the rising waters levelled off, leaving the Florida Keys, mainland, and Biscayne Bay with something similar to their current appearance.3 The keys change. -
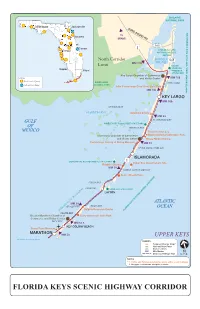
FL Keys Application Map 11X17.Ai
BISCAYNE NATIONAL PARK Tallahassee Jacksonville CARD SOUND RD To Daytona MIAMI Orlando 905 Cocoa 1 Tampa CROCODILE LAKE NATIONAL WILDLIFE REFUGE North Corridor BARNES 905 MM 110 SOUND Limit DAGNY Naples JOHNSON Miami HAMMOCK STATE PARK Key Largo Chamber of Commerce LEGEND and Visitor Center MM 106 Florida Scenic Highway EVERGLADES KEY LARGO National Scenic Byway NATIONAL PARK John Pennekamp Coral Reef State Park MM 102 JOHN PENNEKAMP CORAL REEF STATE PARK (UNDERWATER) KEY LARGO MM 100 OYSTER KEYS FLORIDA BAY Wild Bird Center MM 93 GULF PLANTATION KEY WINDLEY KEY FOSSIL REEF STATE PARK OF WINDLEY KEY MEXICO Theater of the Sea Islamorada Chamber of Commerce Marine Mammal Adventure Park and Visitor Center Whale Harbor Marina Florida Keys History of Diving Museum MM 83 UPPER MATECUMBE KEY ISLAMORADA LIGNUMVITAE KEY BOTANICAL STATE PARK 1 Indian Key State Historic Site Robbie’s Marina MM 78 LOWER MATECUMBE KEY Anne’s Beach Park ARY FIESTA KEY LONG KEY LONG KEY STATE PARK LAYTON ATLANTIC MM 59 GRASSY KEY DUCK KEY OCEAN Dolphin Research Center INTRACOASTAL WATERWAY FLORIDA KEYS NATIONAL MARINE SANCTU CRAWL KEY Greater Marathon Chamber of Curry Hammock State Park Commerce and Visitor Center VACA KEY MM 53.5 KEY COLONY BEACH Crane Point Museum MARATHON MM 50 UPPER KEYS continued on next page LEGEND xxx Features/Itinerary Stops* xxx National/State Parks xxx Visitor Centers MM Mile Marker Overseas Heritage Trail NOTES * 1. Diving and Fishing opportunities along entire scenic highway. 2. No gaps or intrusions along the corridor. FLORIDA KEYS SCENIC -

Archaeological Investigations on the Upper Florida Keys by JOHN M
Archaeological Investigations On The Upper Florida Keys by JOHN M. GOGGIN D URING THE SUMMER of 1940 an archaeological survey was made of a portion of the Upper Florida Keys.' The area covered was from Virginia and Biscayne Keys on the north to Lower Mate- cumbe Key on the south, plus a portion of the west shore of Biscayne Bay. Summer is the least desirable time to attempt work in this area, but circumstances offered no other choice. The area in question was inhabited by the Tekesta* Indians.2 Ap- parently this tribe, like various others in the state, was more or less a political confederation of small local groups. There are many references to the presence of Indians on the Keys or "Los Martires" as the Spanish frequently called them, but few villages are named. Fontaneda does mention some towns, but they appear to have been located on the Lower Keys. In 1607 Governor Ibarra states that he received a visit from some chiefs, among whom were " . .. principal lords of the mouth of Miguel Mora." According to Swanton ".. this name was given to the opening between the Florida Mainland and the Keys on the eastern side." 3 This may be the present Card Sound. The one tribe or village that is men- tioned rather commonly is Matecumbe. This was probably located on the present Lower Matecumbe Key, because on many early maps it is called Old Matecumbe to differentiate it from Upper Matecumbe Key. The earliest mention is in 1573, when the Matecumbe Indians attained notice by killing a number of Spaniards.4 It was again mentioned about 1. -

Marathon Flights Key West Flights
2]DUN 7LIWRQ (OED (YHUJUHHQ %ODNHO\ 'DULHQ FLORIDA $QGDOXVLD (QWHUSULVH 1HZWRQ %ODFNVKHDU LEGEND 3HDUVRQ &DPLOOD 'RWKDQ :D\FURVV 6WDWH&DSLWDO &ROTXLWW 1DVKYLOOH 1DKXQWD 0RXOWULH \ %UXQVZLFN %UHZWRQ Conecuh $GHO National RSXODWLRQ &LWLHVE Forest 3 'RQDOVRQYLOOH *HQHYD /DNHODQG +RPHUYLOOH $/$%$0$ ,QWHUVWDWH 86 6WDWH&RXQW\ %UDWW 1RPD &HQWXU\ Blackwater River 3D[WRQ *DVNLQ &DPSEHOOWRQ :RRGELQH 2DN*URYH State Forest /DXUHO *UDFHYLOOH 0DORQH -D\ %ODFNPDQ %HUU\GDOH +LOO /HRQLD + 2 / 0 ( 6 -DFRE ( 6 :DOQXW ,]DJRUD )UHHZD\ 7ROO 0XQVRQ &LW\ *UHHQZRRG %DLQEULGJH &DLUR Cumberland Island %D\ +LOO 0F'DYLG &DPSWRQ 3URVSHULW\ *OHQGDOH 'HOOZRRG 7KRPDVYLOOH National Seashore 6DUDODQG 0LQHWWH & $ 0 % , $ Escambia 9DOGRVWD )RONVWRQ )UHHZD\ %RQLID\ &RWWRQGDOH River %DNHU .LQJVODQG 3ULQFLSOH+Z\ 6 $ 1 7 $ &UHVWYLHZ 'H)XQLDN &KLSOH\ 4XLWPDQ 3ULFKDUG 0DULDQQD Lake %RXORJQH 5 2 6 $ Okefenokee 6DLQW0DU\V 0ROLQR +ROW 6SULQJV Seminole 0LQRU+Z\ Wilderness 1 $ 6 6 $ 8 3DVVHQJHU5DLO 2 . $ / 2 2 6 $ 6QHDGV &KDWWDKRRFKHH +LOOLDUG %HFNHU 0RELOH : $ / 7 2 1 *(25*,$ )HUQDQGLQD - $ & . 6 2 1 / ( 2 1 <XOHH &RWWDJH+LOO +DUROG : $ 6 + , 1 * 7 2 1 5RXQG &RXQW\%RXQGDU\ 0LOWRQ 6LQN&UHHN *UHWQD +DYDQD %HDFK 'DSKQH Eglin Air 9HUQRQ /DNH )/25,'$ -HQQLQJV 6WDWH%RXQGDU\ 3DFH 0LFFRVXNHH 3LQHWWD $PHOLD&LW\ *RQ]DOH] Force Base $OWKD *UHHQVERUR 4XLQF\ 0RQWLFHOOR &DOODKDQ Torreya +$0,/721 Timucuan Ecological (QVOH\ 1LFHYLOOH 3RUWODQG Choctawhatchee %UDGIRUGYLOOH 8UEDQ 3DUN)RUHVW 0LOLWDU\ )DLUKRSH )HUU\3DVV 9DOSDUDLVR State Park 0LGZD\ John M. Bethea And Historic Preserve )UHHSRUW River 'ULIWRQ -DVSHU %HOOYLHZ Escambia 0DGLVRQ State Forest 5DWOLII * $ ' 6 ' ( 1 $$ Bay +ROOH\ :ULJKW %ORXQWVWRZQ *UHHQYLOOH 7D\ORU Choctawhatchee Bay %ULVWRO /HH Little Talbot Island :HVW3HQVDFROD 0DU\(VWKHU 2FHDQ&LW\ (EUR 7DOODKDVVHH State Park 3HQVDFROD 6DQWD5RVD *HQRD $ & $ / + 2 8 1 (OODYLOOH % $ . -

Biscayne National Park: General Management Plan; P
B ISCAYNE NATIONAL PARK H ISTORIC RESOURCE STUDY January 1998 Jennifer Brown Leynes and David Cullison National Park Service Southeast Region Atlanta, Georgia CONTENTS Figure Credits iv List of Figures v Foreword vii Chapter One: Introduction 1 Chapter Two: Background History 7 Chapter Three: Recreational Development of Miami and Biscayne Bay, 1896-1945 19 Associated Properties 32 Registration Requirements/Integrity 36 Contributing Properties 37 Noncontributing Properties 37 Chapter Four: Management Recommendations 39 Bibliography 41 Appendix A: Architectural Descriptions and Recommended Treatments for the Honeywell Complex on Boca Chita Key A-1 Appendix B: August Geiger B-1 Appendix C: Cleaning Stains on Historic Stone Masonry C-1 Appendix D: Fowey Rocks Lighthouse D-1 Appendix E: Historic Base Map E-1 Appendix F: National Register Nomination for Boca Chita Key Historic District F-1 Index G-1 iii FIGURE CREDITS Cover: Jim Adams for National Park Service; p. 2: National Park Service, Biscayne National Park: General Management Plan; p. 4: Biscayne National Park archives; p. 8: National Park Service, Biscayne National Park: General Management Plan; pp. 12, 15, 20, 23: Historical Museum of Southern Florida, pp. 26, 28-31: Biscayne National Park archives; p. 33: Jim Adams for National Park Service; p. 34: Biscayne National Park archives; p. A-7: Jim Adams for National Park Service; pp. A-10, D-1: David Cullison for National Park Service. iv FIGURES Figure 1. Location of Biscayne National Park 2 Figure 2. Aerial view of Boca Chita Key 4 Figure 3. Biscayne National Park boundaries 8 Figure 4. “Wreckers at Work” from Harper’s New Monthly Magazine, April 1859 12 Figure 5. -

REPUBLICAN Gazette by Republicans, for Republicans
UPPER KEYS REPUBLICAN Gazette By Republicans, For Republicans Vol. 1, No. 3 www.keysgop.com/upper-keys August 2020 Primary races August 18 - please vote! CANDIDATES INSIDE The Upper Keys Republican Club is 250 words. The article submissions pleased to present our third edition of are unedited for content. The Upper the Upper Keys Republican Gazette. Keys Republican Club does not This third edition is a continuation of endorse candidates in primary races. our Candidates Series leading into BACHELER, pg. 3 BELL-THOMPSON, p. 3 the Primary elections. In this edition, No candidate was excluded by the we asked Candidates to tell us why Upper Keys Republican Club. they are running in approximately MONROE COUNTY COMMISSION DAVIS, pg. 3 FORSTER, pg. 2 DISTRICT ONE I was born and raised in Key West. I understand the reasons we live in the Keys and I understand our economy. My wife Cheryl and I have been married for over 48 years. We raised three daughters here HAMMOND, pg. 6 HASKINS, pg. 5 and our granddaughter is growing up here. We spend our time supporting numerous local non-profit organizations, actively serving on many boards throughout Key West and the Keys. We truly enjoy our community and diligently support all the good work that LAVIN, pg. 3 MAJESKA, pg. 2 Craig Cates goes on here. I started and owned several businesses in the Keys, and I understand the difficulties business owners face every day. As Mayor, I had an open-door policy. I focused on the city’s budget and keeping our community affordable for its residents while balancing quality of life with our thriving tourism industry. -

Florida Keys Sea Heritage Journal
Florida Keys Sea Heritage Journal VOL. 21 NO. 4 Summer 2011 USS SHARK OFFICIAL QUARTERLY PUBLICATION OF THE KEY WEST MARITIME HISTORICAL SOCIETY Oil and Gas Exploration in the Florida Keys By J. Gregory Griffin Copyright 2011 In 1938 Thomas Kenny had a contract for bridge and road repair on Bahia Honda Key. In the process of blasting in preparation for leveling the road, a large piece of rock was thrown out of the ground, and the excavation was almost immediately filled with oil. He reported his findings to the Overseas Road and Toll Bridge District Commission on January eighth. An examination of the land was made and compared to the soil found in the oil producing areas in Texas and Louisiana. Extensive surveys, geologists, and seismic tests followed with great expectation. This was the first direct evidence of the possibility of petroleum in the Florida Keys. Subsequently, fourteen exploratory oil wells were drilled in the Keys beginning around the time oil was discovered at the Sunniland Field in 1943 in Collier County. The Second World War ended in 1945 with the total victory of the Allies over the Axis powers Big Pine Key oil well in 1959. Photo credit: Pat Parks. of Germany and Japan. Oil, in all its forms, was an indispensable TNT) for bombs, synthetic rubber War could not have been won product to the Allied forces for tires, gasoline (particularly at without plentiful quantities of oil. around the world. Refinement of 100-octane levels) for trucks, tanks, With peace came the desire oil yielded tarmac for runways, jeeps, and airplanes, and lubricant toluene (the chief component of oil for guns and machinery. -

Fkaa Bacteria Monthly Report
FKAA BACTERIA MONTHLY REPORT PWSID# 4134357 Month: December 2017 H.R.S. LAB # E56717 & E55757 MMO-MUG/ 100ML CL2 PH RETEST MMO-MUG/ DATE 100ML CL2 PH SERVICE AREA # 1 S.I. LAB Date Sampled 12/05/17 114 US Navy Trumbo Pt. OMI Fleming Key Bridge A 3.0 9.28 116 827 Eisenhower Drive, Key West A 3.3 9.18 140 Sybtopic Dive Shop 1601 N Roosevelt Blvd A 3.4 9.26 138 Dion's Quick Mart-1000 White St A 3.2 9.32 142 Banana Bay Resort -2319 N Roosevelt Blvd A 3.5 9.33 118 Fairfield Inn-2400 N Roosevelt Blvd A 3.4 9.23 120 US Navy Housing Elem.School-Felton Rd A 3.2 9.32 132 Almond Tree-512 Truman Ave A 3.6 9.33 108 Douglas Square Community Center-800 Emma St A 3.8 9.33 150 Truman Annex-Patio Beach Bldg #4214 A 3.2 9.24 134 Court House-302 Fleming St A 3.0 9.25 110 Guy Havery's-515 Green St A 3.8 9.36 112 Key West Seaport-631 Greene St A 3.5 9.32 136 Land's End Village -#2 William St A 3.8 9.37 152 Navy Medical Hospital-South Roosevelt (A1A-MM4) A 3.9 9.47 102 Sheraton Suites-2001 S Roosvelt Blvd A 3.2 9.24 126 Key West by the Seas Condo-2601 S Roosevelt Blvd A 3.2 9.19 128 1800 Atlantic Condo-1800 Atlantic Blvd A 3.2 9.24 104 1310 Johnson St Key West A 3.2 9.29 106 Santa Maria Joint Venture-1401 Simonton St Key West A 3.2 9.27 130 The Reach Resort-1435 Simonton St Key West A 3.2 9.27 144 Conch Scoops-3214 N Roosevelt Blvd A 3.4 9.25 124 Dr.