REGISTRY Database Summary Sheet (DBSS)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Conformations of Cyclopentasilane Stereoisomers Control Molecular Junction Conductance
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2017 Supporting Information Conformations of Cyclopentasilane Stereoisomers Control Molecular Junction Conductance Haixing Li,1 Marc H. Garner,2 Zhichun Shangguan,3 Timothy A. Su,4 Madhav Neupane,4 Panpan Li,3 Alexandra Velian,4 Michael L. Steigerwald,4 Shengxiong Xiao,*3 Colin Nuckolls,*3,4 Gemma C. Solomon,*2 Latha Venkataraman*1,4 1 Department of Applied Physics and Applied Mathematics, Columbia University, New York 10027, USA 2 Nano-Science Center and Department of Chemistry, University of Copenhagen, Universitetsparken 5, 2100 Copenhagen Ø, Denmark 3 The Education Ministry Key Lab of Resource Chemistry, Shanghai Key Laboratory of Rare Earth Functional Materials, Optoelectronic Nano Materials and Devices Institute, Department of Chemistry, Shanghai Normal University, Shanghai 200234, China 4 Department of Chemistry, Columbia University, New York 10027 This version of the ESI replaces the previous version that was published on 13th June 2016. Some references in Part IV on Page 8 and 21 are corrected and updated. 1 Table of Contents Table of Contents .............................................................................................. 2 I. Synthetic procedures and characterization of compounds ......................... 4 II. STM-Break Junction experiment details ..................................................... 6 III. Additional Data ......................................................................................... -

Retention Indices for Frequently Reported Compounds of Plant Essential Oils
Retention Indices for Frequently Reported Compounds of Plant Essential Oils V. I. Babushok,a) P. J. Linstrom, and I. G. Zenkevichb) National Institute of Standards and Technology, Gaithersburg, Maryland 20899, USA (Received 1 August 2011; accepted 27 September 2011; published online 29 November 2011) Gas chromatographic retention indices were evaluated for 505 frequently reported plant essential oil components using a large retention index database. Retention data are presented for three types of commonly used stationary phases: dimethyl silicone (nonpolar), dimethyl sili- cone with 5% phenyl groups (slightly polar), and polyethylene glycol (polar) stationary phases. The evaluations are based on the treatment of multiple measurements with the number of data records ranging from about 5 to 800 per compound. Data analysis was limited to temperature programmed conditions. The data reported include the average and median values of retention index with standard deviations and confidence intervals. VC 2011 by the U.S. Secretary of Commerce on behalf of the United States. All rights reserved. [doi:10.1063/1.3653552] Key words: essential oils; gas chromatography; Kova´ts indices; linear indices; retention indices; identification; flavor; olfaction. CONTENTS 1. Introduction The practical applications of plant essential oils are very 1. Introduction................................ 1 diverse. They are used for the production of food, drugs, per- fumes, aromatherapy, and many other applications.1–4 The 2. Retention Indices ........................... 2 need for identification of essential oil components ranges 3. Retention Data Presentation and Discussion . 2 from product quality control to basic research. The identifi- 4. Summary.................................. 45 cation of unknown compounds remains a complex problem, in spite of great progress made in analytical techniques over 5. -

CAS REGISTRY: Finding CAS Registry Numbers
STN® Quick Reference Card CAS REGISTRYSM: Finding CAS Registry Numbers® When you know the Substance Name Field Use Example /CN Use when you know the complete => E BENZOIC ACID/CN Chemical substance name. EXPAND first to => E “BICYCLO(2.2.1)HEPTANE”/CN Name determine if the name is in the database. Basic Index Use when you know segments of the => S FLUOROMETHYL name or don’t know how the name => S “2,2’” (W) BIPYRID? segments go together. => S PYRIDINE (XW) DICARBOXY? /CNS Use when you want to search for a => S ?MYCIN?/CNS Chemical Name character string embedded in a trade => S ?QUAT/CNS Segments name. Use left and right truncation to search for an embedded character string. Use when you want to require that a => S CHLOROPHENYL/CNS name segment is not part of a larger => S HEXANEDIOIC (XW) segment. DIMETHYL/CNS /HP Use when you want to restrict the search => S 1-PROPANOL/HP Heading to a Heading Parent from a CA index => S 2-PYRIDINECARBO?/HP Parent name. /INS.HP Use when you want to restrict the search => S (PYRIDINE (XW) Index Name to name segments from the Heading CARBONITRIL?)/INS.HP part of a CA index name. Segments - Parent => S FLUOROMETHYL/INS.HP Heading Parent /INS.NHP Use when you want to restrict the search => S MORPHOL?/INS.NHP Index Name to name segments from the non- => S FLUOROMETHYL/INS.NHP Segments - part of a CA index Heading-Parent Non-Heading name. Parent /ONS Use when you want to restrict the search => S VINCAM?/ONS Other Name to name segments from names other => S (INDOLE(XW)AMIN?)/ONS Segments than CA index names such as semi- systematic names, trade names, common names, etc. -

Zr-Modified Zno for the Selective Oxidation of Cinnamaldehyde to Benzaldehyde
catalysts Article Zr-Modified ZnO for the Selective Oxidation of Cinnamaldehyde to Benzaldehyde Pengju Du 1, Tongming Su 1, Xuan Luo 1, Xinling Xie 1, Zuzeng Qin 1,* and Hongbing Ji 1,2 1 School of Chemistry and Chemical Engineering, Guangxi University, Nanning 530004, China 2 School of Chemistry, Sun Yat-sen University, Guangzhou 510275, China * Correspondence: [email protected]; Tel.: +86-771-323-3718 Received: 31 July 2019; Accepted: 21 August 2019; Published: 25 August 2019 Abstract: ZnO and Zr-modified ZnO were prepared using a precipitation method and used for the selective oxidation of cinnamaldehyde to benzaldehyde in the present study. The results showed that physicochemical properties of ZnO were significantly affected by the calcination temperature, and calcination of ZnO at 400 ◦C demonstrated the optimum catalytic activity for the selective oxidation of cinnamaldehyde to benzaldehyde. With 0.01 g ZnO calcined at 400 ◦C for 2 h as a catalyst, 8.0 g ethanol and 2.0 g cinnamaldehyde reacted at an oxygen pressure of 1.0 MPa and 70 ◦C for 60 min, resulting in benzaldehyde selectivity of 69.2% and cinnamaldehyde conversion of 16.1%. Zr was the optimal modifier for ZnO: when Zr-modified ZnO was used as the catalyst, benzaldehyde selectivity reached 86.2%, and cinnamaldehyde conversion was 17.6%. The X-ray diffractometer and N2 adsorption–desorption characterization indicated that doping with Zr could reduce the crystallite size of ZnO (101) and increase the specific surface area of the catalyst, which provided more active sites for the reaction. X-ray photoelectron spectrometer results showed that Zr-doping could exchange the electrons with ZnO and reduce the electron density in the outer layer of Zn, which would further affect benzaldehyde selectivity. -

Gas-Phase Chemistry of Methyl-Substituted Silanes in a Hot-Wire Chemical Vapour Deposition Process
University of Calgary PRISM: University of Calgary's Digital Repository Graduate Studies The Vault: Electronic Theses and Dissertations 2013-08-27 Gas-phase Chemistry of Methyl-Substituted Silanes in a Hot-wire Chemical Vapour Deposition Process Toukabri, Rim Toukabri, R. (2013). Gas-phase Chemistry of Methyl-Substituted Silanes in a Hot-wire Chemical Vapour Deposition Process (Unpublished doctoral thesis). University of Calgary, Calgary, AB. doi:10.11575/PRISM/26257 http://hdl.handle.net/11023/891 doctoral thesis University of Calgary graduate students retain copyright ownership and moral rights for their thesis. You may use this material in any way that is permitted by the Copyright Act or through licensing that has been assigned to the document. For uses that are not allowable under copyright legislation or licensing, you are required to seek permission. Downloaded from PRISM: https://prism.ucalgary.ca UNIVERSITY OF CALGARY Gas-phase Chemistry of Methyl-Substituted Silanes in a Hot-wire Chemical Vapour Deposition Process by Rim Toukabri A THESIS SUBMITTED TO THE FACULTY OF GRADUATE STUDIES IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY DEPARTMENT OF CHEMISTRY CALGARY, ALBERTA August, 2013 © Rim Toukabri 2013 Abstract The primary decomposition and secondary gas-phase reactions of methyl- substituted silane molecules, including monomethylsilane (MMS), dimethylsilane (DMS), trimethylsilane (TriMS) and tetramethylsilane (TMS), in hot-wire chemical vapour deposition (HWCVD) processes have been studied using laser ionization methods in combination with time of flight mass spectrometry (TOF-MS). For all four molecules, methyl radical formation and hydrogen molecule formation have been found to be the common decomposition steps on both tungsten (W) and tantalum (Ta) filaments. -

New Insights Into the Mechanism of Schiff Base Synthesis from Aromatic
A peer-reviewed version of this preprint was published in PeerJ on 15 December 2020. View the peer-reviewed version (peerj.com/articles/ochem-4), which is the preferred citable publication unless you specifically need to cite this preprint. Silva PJ. 2020. New insights into the mechanism of Schiff base synthesis from aromatic amines in the absence of acid catalyst or polar solvents. PeerJ Organic Chemistry 2:e4 https://doi.org/10.7717/peerj-ochem.4 New insights into the mechanism of Schiff base synthesis from aromatic amines in the absence of acid catalyst or polar solvents Pedro J Silva Corresp. 1 1 FP-ENAS/Fac. de Ciências da Saúde, Universidade Fernando Pessoa, Porto, Portugal Corresponding Author: Pedro J Silva Email address: [email protected] Extensive computational studies of the imine synthesis from amines and aldehydes in water have shown that the large-scale structure of water is needed to afford appropriate charge delocalisation and enable sufficient transition state stabilisation. These insights cannot, however, be applied to the understanding of the reaction pathway in apolar solvents due their inability to form extensive hidrogen-bonding networks. In this work, we perform the first computational studies of this reaction in apolar conditions. This density- functional study of the reaction of benzaldehyde with four closely related aromatic amines (aniline, o-toluidine, m-toluidine and p-toluidine) shows that an additional molecule of amine may provide enough stabilization of the first transition state even in the absence of a hydrogen bonding network. Our computations also show that the second reaction step cannot take place unless an extra proton is added to the system but, crucially, that reaction rate is so high that even picomolar amounts of protonated base are enough to achieve realistic rates. -

Synthesis of Aniline and Benzaldehyde Derivatives From
Electronic Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2011 Supporting Information Fe2O3-supported nano-gold catalyzed one-pot synthesis of N-alkylated anilines from nitroarenes and alcohols * Qiling Peng, Yan Zhang, Feng Shi , Youquan Deng Centre for Green Chemistry and Catalysis, Lanzhou Institute of Chemical Physics, Chinese Academy of Sciences, Lanzhou, 730000, China *corresponding author: Tel.: +86-931-4968142; Fax: +86-931-4968116; E-mail: [email protected] 1. Materials and Methods 2. Chlorine concentration measurement 3. Catalyst preparation 4. Table S2. ICP-AES and BET data of various catalysts 5. General procedure for the reaction of nitrobenzene with d7-benzyl alcohol 6. General procedure for the H-D exchange reaction between aniline and D2O 7. Fig. S2. XRD patterns of the catalysts in the experiments. 8. XPS spectra of catalyst samples 9. HRTEM images of the catalyst samples 10.GC-MS spectra of compounds observed by GC-MS 11. Compounds characterization and NMR spectra Electronic Supplementary Material (ESI) for Chemical Communications This journal is © The Royal Society of Chemistry 2011 1. Materials and Methods All solvent and chemicals were obtained commercially and were used as received. NMR spectra were measured using a Bruker ARX 400 or ARX 100 spectrometer at 400 MHz (1H) and 100 MHz 13 ( C). All spectra were recorded in CDCl3 and chemical shifts (δ) are reported in ppm relative to tetramethylsilane referenced to the residual solvent peaks. Mass spectra were in general recorded on an HP 6890/5973 GC-MS. High-resolution TEM analysis was carried out on a JEM 2010 operating at 200 KeV. -

Chemical Name Federal P Code CAS Registry Number Acutely
Acutely / Extremely Hazardous Waste List Federal P CAS Registry Acutely / Extremely Chemical Name Code Number Hazardous 4,7-Methano-1H-indene, 1,4,5,6,7,8,8-heptachloro-3a,4,7,7a-tetrahydro- P059 76-44-8 Acutely Hazardous 6,9-Methano-2,4,3-benzodioxathiepin, 6,7,8,9,10,10- hexachloro-1,5,5a,6,9,9a-hexahydro-, 3-oxide P050 115-29-7 Acutely Hazardous Methanimidamide, N,N-dimethyl-N'-[2-methyl-4-[[(methylamino)carbonyl]oxy]phenyl]- P197 17702-57-7 Acutely Hazardous 1-(o-Chlorophenyl)thiourea P026 5344-82-1 Acutely Hazardous 1-(o-Chlorophenyl)thiourea 5344-82-1 Extremely Hazardous 1,1,1-Trichloro-2, -bis(p-methoxyphenyl)ethane Extremely Hazardous 1,1a,2,2,3,3a,4,5,5,5a,5b,6-Dodecachlorooctahydro-1,3,4-metheno-1H-cyclobuta (cd) pentalene, Dechlorane Extremely Hazardous 1,1a,3,3a,4,5,5,5a,5b,6-Decachloro--octahydro-1,2,4-metheno-2H-cyclobuta (cd) pentalen-2- one, chlorecone Extremely Hazardous 1,1-Dimethylhydrazine 57-14-7 Extremely Hazardous 1,2,3,4,10,10-Hexachloro-6,7-epoxy-1,4,4,4a,5,6,7,8,8a-octahydro-1,4-endo-endo-5,8- dimethanonaph-thalene Extremely Hazardous 1,2,3-Propanetriol, trinitrate P081 55-63-0 Acutely Hazardous 1,2,3-Propanetriol, trinitrate 55-63-0 Extremely Hazardous 1,2,4,5,6,7,8,8-Octachloro-4,7-methano-3a,4,7,7a-tetra- hydro- indane Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]- 51-43-4 Extremely Hazardous 1,2-Benzenediol, 4-[1-hydroxy-2-(methylamino)ethyl]-, P042 51-43-4 Acutely Hazardous 1,2-Dibromo-3-chloropropane 96-12-8 Extremely Hazardous 1,2-Propylenimine P067 75-55-8 Acutely Hazardous 1,2-Propylenimine 75-55-8 Extremely Hazardous 1,3,4,5,6,7,8,8-Octachloro-1,3,3a,4,7,7a-hexahydro-4,7-methanoisobenzofuran Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime 26419-73-8 Extremely Hazardous 1,3-Dithiolane-2-carboxaldehyde, 2,4-dimethyl-, O- [(methylamino)-carbonyl]oxime. -

Grignard Reagents and Silanes
GRIGNARD REAGENTS AND SILANES By B. Arkles REPRINTED FROM HANDBOOK OF GRIGNARD REAGENTS by G. Silverman and P. Rakita Pages 667-675 Marcel Dekker, 1996 Gelest, Inc. 612 William Leigh Drive Tullytown, Pa. 19007-6308 Phone: [215] 547-1015 Fax: [215] 547-2484 Grignard Reagents and Silanes 32 Grignard Reagents and Silanes BARRY ARKLES Gelest Inc., Tullytown, Pennsylvania I. INTRODUCTION This review considers two aspects of the interaction of Grignards with silanes. First, focusing on technologies that are still viable within the context of current organosilane and silicone technology, guidelines are provided for silicon-carbon bond formation using Grignard chemistry. Second, the use of silane-blocking agents and their stability in the presence of Grignard reagents employed in organic synthesis is discussed. II. FORMATION OF THE SILICON-CARBON BOND A. Background The genesis of current silane and silicone technology traces back to the Grignard reaction. The first practical synthesis of organosilanes was accomplished by F. Stanley Kipping in 1904 by the Grignard reaction for the formation of the silicon-carbon bond [1]. In an effort totaling 57 papers, he created the basis of modern organosilane chemistry. The development of silicones by Frank Hyde at Corning was based on the hydrolysis of Grignard-derived organosilanes [2]. Dow Corning, the largest manufacturer of silanes and silicones, was formed as a joint venture between Corning Glass, which had silicone product technology, and Dow, which had magnesium and Grignard technology, during World War II. In excess of 10,000 silicon compounds have been synthesized by Grignard reactions. Ironically, despite the versatility of Grignard chemistry for the formation of silicon-carbon bonds, its use in current silane and silicone technology has been supplanted by more efficient and selective processes for the formation of the silicon-carbon bond, notably by the direct process and hydrosilylation reactions. -

Reactions of Aromatic Compounds Just Like an Alkene, Benzene Has Clouds of Electrons Above and Below Its Sigma Bond Framework
Reactions of Aromatic Compounds Just like an alkene, benzene has clouds of electrons above and below its sigma bond framework. Although the electrons are in a stable aromatic system, they are still available for reaction with strong electrophiles. This generates a carbocation which is resonance stabilized (but not aromatic). This cation is called a sigma complex because the electrophile is joined to the benzene ring through a new sigma bond. The sigma complex (also called an arenium ion) is not aromatic since it contains an sp3 carbon (which disrupts the required loop of p orbitals). Ch17 Reactions of Aromatic Compounds (landscape).docx Page1 The loss of aromaticity required to form the sigma complex explains the highly endothermic nature of the first step. (That is why we require strong electrophiles for reaction). The sigma complex wishes to regain its aromaticity, and it may do so by either a reversal of the first step (i.e. regenerate the starting material) or by loss of the proton on the sp3 carbon (leading to a substitution product). When a reaction proceeds this way, it is electrophilic aromatic substitution. There are a wide variety of electrophiles that can be introduced into a benzene ring in this way, and so electrophilic aromatic substitution is a very important method for the synthesis of substituted aromatic compounds. Ch17 Reactions of Aromatic Compounds (landscape).docx Page2 Bromination of Benzene Bromination follows the same general mechanism for the electrophilic aromatic substitution (EAS). Bromine itself is not electrophilic enough to react with benzene. But the addition of a strong Lewis acid (electron pair acceptor), such as FeBr3, catalyses the reaction, and leads to the substitution product. -

Synthesis of Some Imines and Investigation of Their Biological Activity
ISSN: 0973-4945; CODEN ECJHAO E-Journal of Chemistry http://www.e-journals.net 2009, 6(3), 629-632 Synthesis of Some Imines and Investigation of their Biological Activity MAJED M. HANIA Chemistry Department, The Islamic University, Gaza P.O.Box 108, Gaza, Palestine. [email protected] Received 12 December 2008; Accepted 5 February 2009 Abstract : Some N-benzylidene aniline derivatives have been synthesized and tested as antibacterial agents. The results of the in vitro tests showed that most of the synthesized compounds were antibacterial inactive against E.coli. Keywords: Synthesis, Imines, Investigation, Antibacterial activity. Introduction Schiff bases such as substituted N-benzylidene aniline are a class of important compounds in medicinal and pharmaceutical field. They show biological activities including antibacterial 1-4, antifungal 5-6, anticancer 7-10 , and herbicidal 11 activities . In view of the above observations, the synthesis of N-benzylidene aniline derivatives have been developed starting from various substituted benzylidene anilines with the aim of investigating their antibacterial activities. Experimental Melting points were determined in open capillaries and are uncorrected. IR spectra were recorded in KBr on Perkin-Elmer 883 spectrometer. The UV spectra were obtained by using UV-vis spectrophotometer instrument, Perkin-Elmer Lambda 20.1 nm. All the compounds gave satisfactory analysis. Benzaldehyde, 4-methylbenzaldehyde, 4-methoxylbenzaldehyde, 4-nitobenzaldehyde and 4-chlorobenzaldehyde were obtained from Sigma- -
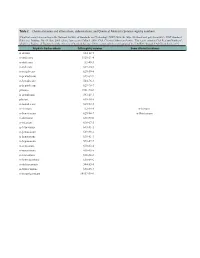
Table 2. Chemical Names and Alternatives, Abbreviations, and Chemical Abstracts Service Registry Numbers
Table 2. Chemical names and alternatives, abbreviations, and Chemical Abstracts Service registry numbers. [Final list compiled according to the National Institute of Standards and Technology (NIST) Web site (http://webbook.nist.gov/chemistry/); NIST Standard Reference Database No. 69, June 2005 release, last accessed May 9, 2008. CAS, Chemical Abstracts Service. This report contains CAS Registry Numbers®, which is a Registered Trademark of the American Chemical Society. CAS recommends the verification of the CASRNs through CAS Client ServicesSM] Aliphatic hydrocarbons CAS registry number Some alternative names n-decane 124-18-5 n-undecane 1120-21-4 n-dodecane 112-40-3 n-tridecane 629-50-5 n-tetradecane 629-59-4 n-pentadecane 629-62-9 n-hexadecane 544-76-3 n-heptadecane 629-78-7 pristane 1921-70-6 n-octadecane 593-45-3 phytane 638-36-8 n-nonadecane 629-92-5 n-eicosane 112-95-8 n-Icosane n-heneicosane 629-94-7 n-Henicosane n-docosane 629-97-0 n-tricosane 638-67-5 n-tetracosane 643-31-1 n-pentacosane 629-99-2 n-hexacosane 630-01-3 n-heptacosane 593-49-7 n-octacosane 630-02-4 n-nonacosane 630-03-5 n-triacontane 638-68-6 n-hentriacontane 630-04-6 n-dotriacontane 544-85-4 n-tritriacontane 630-05-7 n-tetratriacontane 14167-59-0 Table 2. Chemical names and alternatives, abbreviations, and Chemical Abstracts Service registry numbers.—Continued [Final list compiled according to the National Institute of Standards and Technology (NIST) Web site (http://webbook.nist.gov/chemistry/); NIST Standard Reference Database No.